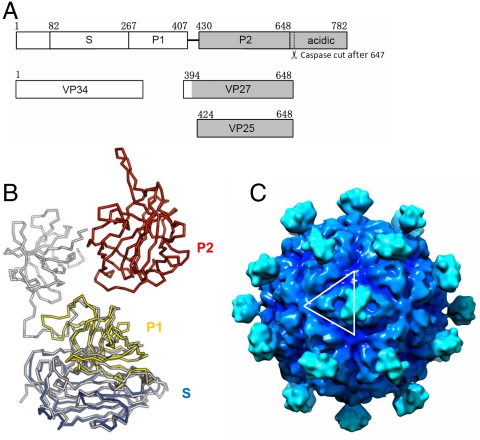
Fig. 3.
Astrovirus capsid structure model. (A) Domain structure of astrovirus CP of serotype 8. Astrovirus CP has a conserved region (in white), a variable region and an acidic C-terminal region (in gray). The S and P1 domain of astrovirus as defined in this paper can be roughly mapped to the conserved region. The P2 domain, however, is mapped to the variable region. The coverage of VP34, VP27, and VP24 is also indicated. Although the N-terminal residues of these three polypeptides have been previously mapped, it is not clear exactly where they end. Our crystal structure of the surface spike strongly argues that the C-terminal residues of both VP27 and VP25 are Arg648, which is the first trypsin cleavage site after the last ordered residues (Pro645) in the spike. This residue is also the only trypsin susceptible site that would explain the size of VP28 and VP27, because these two proteins have the same N-terminal residue (12). The C terminus of VP34 remains ambiguous. (B) Astrovirus CP superimposed onto HEV CP. HEV CP is shown in gray, whereas the astrovirus CP is colored by domains with S in blue, P1 in yellow, and P2 in red. (C) T = 3 astrovirus capsid model rendered at 15-Å resolution. The white lines highlight one asymmetric unit. The contour level was chosen so that the volume is consistent with 10 Å3 per non-hydrogen atom. The isosurfaces were drawn using Chimera (40).