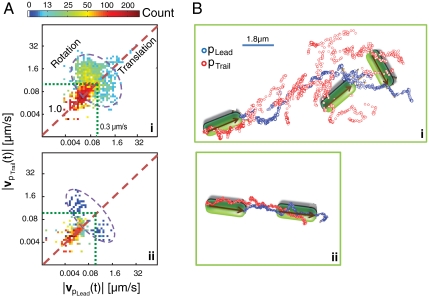
Fig. 2.
Identical ΔfliM bacteria exhibit individuation of motion preferences. (A) 2D histogram of |vpLead(t)| versus |vpTrail(t)|, from datasets of two different ΔfliM mutant bacteria. The dashed line (slope = 1.0) is a guide to the eye to indicate translational motion (|vpLead(t)| = |vpTrail(t)|); the dashed circle is a guide to the eye to indicate rotational motion (|vpLead(t)| ≠ |vpTrail(t)|); the dotted green lines indicate the velocity threshold (|vpLead,pTrail| = 0.3 μm/s) separating release actions and pull actions. Genetically identical bacteria exhibit distinct individual motion preferences: (i) rotation; (ii) translation. (B) Portion of trajectories from (top) bacterium (i), with schematic illustrating rotational motion, and (bottom) bacterium (ii), with schematic illustrating translational motion. In (B) the blue and red circles indicate positions of pLead and pTrail, respectively.