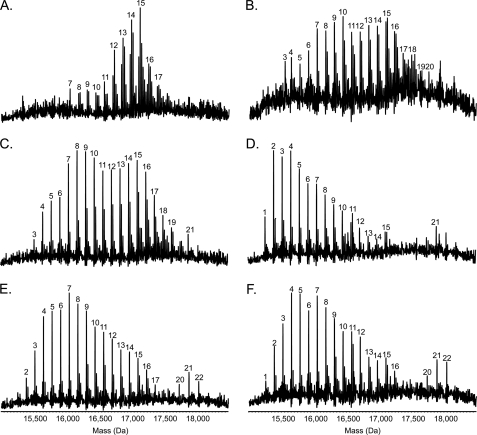
FIGURE 5.
Mass spectrometry analysis of intact pilins. Pili were isolated and subjected to electrospray ionization-MS as described under “Experimental Procedures” to determine the extent and pattern of pilin glycosylation. Presented here are the reconstructed molecular mass profiles. A, Pa5196 6250–51 double mutant. B, PAO1 NP + AWX + 6245–6251. C, PAO1 NP + AWX + 6245–6249. D, PAO1 NP + AWX + 6246–6249. E, PAO1 NP + AWX + 6245–6249, 6247::GmFRT. F, PAO1 NP + AWX + 6246–6249, 6247::GmFRT. As previously observed for the Pa5196 wild type (3), a characteristic pattern of evenly spaced peaks, each separated by 132 Da (the mass of a single arabinofuranose unit), is observed in the reconstructed molecular mass profiles of all the strains analyzed. Sodium adduct peaks are prominent in the profiles of some of the isolates. The mass of the unmodified protein is 15,132 Da; the peaks are labeled with the number of d-Araf residues present on the pilin.