β-Galactosidase from the psychrotrophic and halotolerant Planococcus sp. L4 (BgaP) was crystallized and a complete data set was collected. There are six protein subunits in the asymmetric unit.
Keywords: cold-active β-galactosidases, Planococcus sp. L4, BgaP
Abstract
β-Galactosidases catalyze the hydrolysis of a galactosyl moiety from the nonreducing termini of oligosaccharides or from glycosides. A novel GH family 42 cold-active β-galactosidase identified from the psychrotrophic and halotolerant Planococcus sp. L4 (BgaP) was crystallized and a complete data set was collected from a single frozen crystal on an in-house X-ray source. The crystal diffracted to 2.8 Å resolution and belonged to space group P1, with unit-cell parameters a = 104.29, b = 118.12, c = 121.12 Å, α = 62.66, β = 69.48, γ = 70.74°. A likely Matthews coefficient of 2.58 Å3 Da−1 and solvent content of 52.32% suggested the presence of six protein subunits in the asymmetric unit.
1. Introduction
β-Galactosidases (β-d-galactoside galactohydrolases; EC 3.2.1.23) catalyse the hydrolysis of the galactosyl moiety from the nonreducing termini of oligosaccharides or from glycosides. These enzymes belong to families 1, 2, 35 and 42 of the glycoside hydrolase classification (GH families 1, 2, 35 and 42; Henrissat & Davies, 1997 ▶) and are widely distributed in microorganisms, plants and animals. β-Galactosidases are particularly useful in industrial biotechnology, e.g. for the hydrolysis of lactose in dairy products for people who are lactose-intolerant and for the production of oligosaccharides and sulfated disaccharides (Gaur et al., 2006 ▶; Benesová et al., 2010 ▶; Rhimi et al., 2010 ▶; Shukla & Wierzbicki, 1975 ▶). A large number of β-galactosidases have been exploited and characterized and some of them have shown potential for industrial application (Ghatak et al., 2010 ▶; Grogan, 1991 ▶; Medow et al., 1990 ▶; Kim et al., 2006 ▶; Nagy et al., 2001 ▶).
Dairy products are usually shipped and stored under refrigerated conditions in order to prevent the growth of bacteria or fungi. While cold-active β-galactosidases would be able to perform the hydrolysis of lactose at the same time as shipping and storage, an extra process to warm dairy products up to ordinary temperature or higher is required for mesophilic or thermophilic β-galactosidases to work functionally. Thus, the use of cold-active β-galactosidases could avoid contamination problems caused by bacteria or fungi and decrease energy consumption, and they have received much attention from the food industry (Karasová-Lipovová et al., 2003 ▶; Skálová et al., 2005 ▶; Siddiqui & Cavicchioli, 2006 ▶; Ferrer et al., 2007 ▶).
BgaP is a novel GH family 42 cold-active β-galactosidase identified from the psychrotrophic and halotolerant Planococcus sp. L4 (Hu et al., 2007 ▶). BgaP exhibits maximal activity at 293 K and pH 6.8 and retains 27% of its activity at 273 K. It is able to hydrolyze 34% of cow milk lactose after 60 min at 278 K. At 293 K, the catalytic efficiency of BgaP for lactose is 47 times higher than that of Escherichia coli β-galactosidase. BgaP has a high tolerance for salt and retains 27 or 22% activity in the presence of 5 M KCl or 5 M NaCl, respectively. In light of these properties, BgaP will be more efficient and useful than mesophilic, thermophilic and other cold-active enzymes in the dairy industry. However, this enzyme is particularly thermolabile, losing all activity after only 10 min at 318 K.
We are especially interested in investigating the structural basis of this cold-active β-galactosidase and its protein engineering. The most similar protein to BgaP in the PDB is a β-galactosidase of GH family 42 from Thermus thermophilus A4 (PDB entry 1kwk; Hidaka et al., 2002 ▶), which shares 32% sequence identity. Very interestingly, the T. thermophilus A4 β-galactosidase is extremely stable under thermophilic conditions and retains full activity after incubation at 343 K for 20 h (Hidaka et al., 2002 ▶). Other β-galactosidases of known three-dimensional structure, including the cold-active β-galactosidase from Arthrobacter sp. C2-2 (PDB entry 1yq2; Skálová et al., 2005 ▶), all have sequence identities of below 16%. An accurate three-dimensional structure of BgaP will provide new insights into its catalytic mechanism and thermostability. This paper presents the initial steps towards the three-dimensional structure determination of BgaP.
2. Materials and methods
2.1. Cloning, expression and purification
Cloning of the BgaP gene has been described previously (Hu et al., 2007 ▶). For crystallization, the whole BgaP gene was subcloned into expression vector pQE-30 between BamHI and HindIII sites, with 12 additional amino acids (MRGSHHHHHHGS–), including a hexahistidine tag, at the N-terminus. E. coli strain M15 (pREP4) (Qiagen) was used for protein expression. The transformant bacteria were grown in LB medium containing 50 µg ml−1 ampicillin at 310 K to an OD600 of 0.8. Expression of the target protein was induced by adding IPTG to a final concentration of 0.1 mM and the cells were grown for a further 20 h at 289 K. The cells were collected by centrifugation and then resuspended in cold lysis buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl and 1 mg ml−1 lysozyme). The suspension was sonicated and the soluble proteins in the supernatant were recovered by centrifugation at 18 000g for 35 min at 277 K. The supernatant was loaded onto an Ni-chelating HisTrap FF column (GE Healthcare, USA) which had been pre-equilibrated with binding buffer (50 mM NaCl, 50 mM Tris, 10 mM imidazole pH 8.0). The column was washed with binding buffer followed by washing buffer (50 mM NaCl, 50 mM Tris, 30 mM imidazole pH 8.0). The recombinant protein was eluted with elution buffer (50 mM NaCl, 50 mM Tris, 250 mM imidazole pH 8.0). The purified protein was filtered and concentrated using Amicon Ultra-15 (Millipore, USA) in crystallization buffer consisting of 50 mM NaCl, 5 mM Tris pH 8.0, 5% glycerol prior to crystallization setup. All column-chromatography and enzyme-concentration steps were performed at 277 K. The affinity tag was not removed. The purity of the protein was examined by 10% SDS–PAGE and was determined to be >98%.
2.2. Crystallization
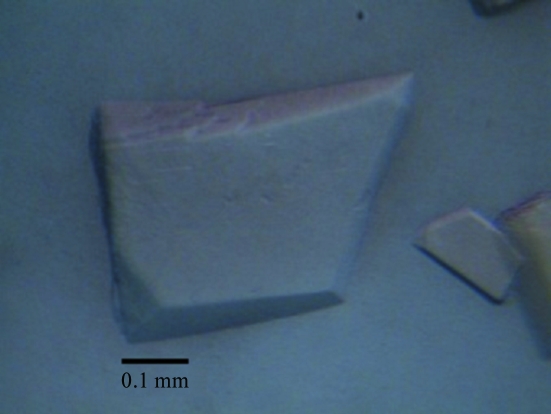
Initial crystallization screening was carried out at 289 K in 24-well plates using the hanging-drop vapour-diffusion method. Each drop consisted of 2 µl protein solution (20 mg ml−1) and 2 µl crystallization solution and was equilibrated against 500 µl crystallization solution. Crystals were observed in less than 1 d using condition No. 26 of Crystal Screen 2 (Hampton Research), which consists of 0.2 M ammonium sulfate, 30% PEG 5000, 0.1 M MES pH 6.5. This condition was optimized by variation of the precipitant, salts, buffer and pH. The optimum condition consisted of 0.2 M ammonium sulfate, 23% PEG 4000, 0.1 M MES pH 6.5. Plate-shaped crystals suitable for diffraction data collection were obtained (Fig. 1 ▶) in 1 d at 289 K.
Figure 1.
Plate-shaped crystal of BgaP with dimensions of 0.10 × 0.30 × 0.40 mm.
2.3. X-ray diffraction data collection
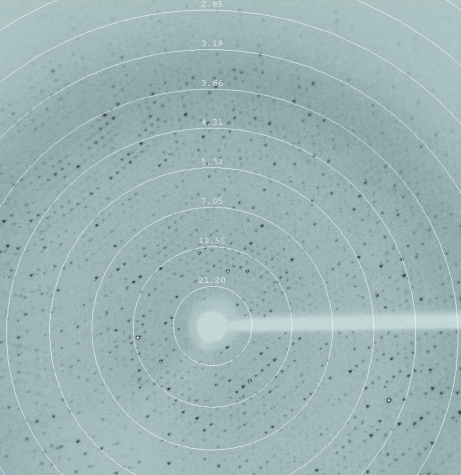
For X-ray diffraction experiments, a crystal was picked up from the crystallization drop using a nylon loop and flash-frozen in a dry nitrogen-gas stream at 100 K. A complete X-ray diffraction data set was collected at a wavelength of 1.5418 Å on our in-house Oxford Diffraction Xcalibur Nova diffractometer operating at 40 kV and 0.8 mA. The exposure time and crystal oscillation angles were set to 50 s and 0.5°, respectively. The crystal-to-detector distance was maintained at 120 mm. The crystal diffracted X-rays to 2.8 Å resolution. A total of 950 images were recorded using a 165 mm Onyx CCD detector. Data sets were processed and scaled using CrysAlisPro (v.1.171.33.49; Oxford Diffraction) and programs from the CCP4 suite (Winn et al., 2011 ▶). A typical diffraction image of the BgaP crystal is shown in Fig. 2 ▶.
Figure 2.
A typical diffraction pattern of a BgaP crystal. The exposure time was 50 s, the crystal-to-detector distance was 120.0 mm and the oscillation range per frame was 0.5°. The diffraction image was collected on a 165 mm Onyx CCD detector.
3. Results and discussion
The concentrated protein was significantly pink in colour and could be crystallized in various forms (plates, rods, needles, hexagons) at 289 or 277 K using a wide range of reservoir solutions comprising 0.2–0.4 M ammonium sulfate, 18–26% PEG (PEG 2000, 3350, 4000, 6000 or 8000), 0.1 M MES pH 6.5 or 0.1 M sodium cacodylate pH 6.5 using the hanging-drop vapour-diffusion method. However, these crystals appeared to be disordered, as shown by their poor diffraction quality. Only by mixing 3 µl protein solution with 3 µl buffer consisting of 0.2 M ammonium sulfate, 23% PEG 4000, 0.1 M MES pH 6.5 and equilibrating against 500 µl reservoir solution (0.2 M ammonium sulfate, 25% PEG 3350, 0.1 M sodium cacodylate pH 6.5) could we obtain good plate-shaped crystals. These crystals appeared in 1 d and diffracted X-rays to 2.8 Å resolution on our in-house X-ray source. The X-ray diffraction data collected from a single crystal were processed using CrysAlisPro (v.1.171.33.49; Oxford Diffraction) and scaled using SCALA from the CCP4 suite (Winn et al., 2011 ▶) at a resolution cutoff of 2.8 Å. The crystal belonged to space group P1, with unit-cell parameters a = 104.29, b = 118.12, c = 121.12 Å, α = 62.66, β = 69.48, γ = 70.74°. The data-collection and processing statistics are summarized in Table 1 ▶. The likely Matthews coefficient (Matthews, 1968 ▶) and solvent content of 2.58 Å3 Da−1 and 52.32%, respectively, suggested the presence of six protein subunits in the asymmetric unit. Our efforts are now aimed at collecting a high-quality data set at a synchrotron station and structure determination is under way by molecular replacement.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Crystallization conditions | 0.2 M ammonium sulfate, 23% PEG 4000, 0.1 M MES pH 6.5 |
| Wavelength (Å) | 1.5418 |
| Temperature (K) | 100 |
| Resolution range (Å) | 24.41–2.80 (2.95–2.80) |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 104.29, b = 118.12, c = 121.11, α = 62.66, β = 69.48, γ = 70.74 |
| Observed reflections | 307989 (26823) |
| Unique reflections | 114916 (16491) |
| Data completeness (%) | 99.1 (96.9) |
| Multiplicity | 2.7 (1.6) |
| 〈I/σ(I)〉 | 9.2 (3.7) |
| Rmerge (%) | 0.085 (0.167) |
| Matthews coefficient (Å3 Da−1) | 2.58 |
| Solvent content (%) | 52.32 |
Acknowledgments
The authors gratefully thank the Natural Science Foundation of China (No. 30800169), the National High Technology Research and Development Program of China (863 Program, 2007AA10Z308) and the Research Fund for the Doctoral Program of Higher Education of China (200805581146).
References
- Benesová, E., Lipovová, P., Dvoráková, H. & Králová, B. (2010). Glycobiology, 20, 442–451. [DOI] [PubMed]
- Ferrer, M., Golyshina, O., Beloqui, A. & Golyshin, P. N. (2007). Curr. Opin. Microbiol. 10, 207–214. [DOI] [PubMed]
- Gaur, R., Pant, H., Jain, R. & Khare, S. K. (2006). Food Chem. 97, 426–430.
- Ghatak, A., Guha, A. K. & Ray, L. (2010). Appl. Biochem. Biotechnol. 162, 1678–1688. [DOI] [PubMed]
- Grogan, D. W. (1991). Appl. Environ. Microbiol. 57, 1644–1649. [DOI] [PMC free article] [PubMed]
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol. 7, 637–644. [DOI] [PubMed]
- Hidaka, M., Fushinobu, S., Ohtsu, N., Motoshima, H., Matsuzawa, H., Shoun, H. & Wakagi, T. (2002). J. Mol. Biol. 322, 79–91. [DOI] [PubMed]
- Hu, J. M., Li, H., Cao, L. X., Wu, P. C., Zhang, C. T., Sang, S. L., Zhang, X. Y., Chen, M. J., Lu, J. Q. & Liu, Y. H. (2007). J. Agric. Food Chem. 55, 2217–2224. [DOI] [PubMed]
- Karasová-Lipovová, P., Strnad, H., Spiwok, V., Malá, Š., Králová, B. & Russell, N. J. (2003). Enzyme Microb. Technol. 33, 836–844.
- Kim, Y.-S., Park, C.-S. & Oh, D.-K. (2006). Enzyme Microb. Technol. 39, 903–908.
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Medow, M. S., Thek, K. D., Newman, L. J., Berezin, S., Glassman, M. S. & Schwarz, S. M. (1990). Am. J. Dis. Child. 144, 1261–1264. [DOI] [PubMed]
- Nagy, Z., Kiss, T., Szentirmai, A. & Biró, S. (2001). Protein Expr. Purif. 21, 24–29. [DOI] [PubMed]
- Rhimi, M., Boisson, A., Dejob, M., Boudebouze, S., Maguin, E., Haser, R. & Aghajari, N. (2010). Res. Microbiol. 161, 515–525. [DOI] [PubMed]
- Shukla, T. P. & Wierzbicki, L. E. (1975). CRC Crit. Rev. Food Sci. Nutr. 5, 325–356.
- Siddiqui, K. S. & Cavicchioli, R. (2006). Annu. Rev. Biochem. 75, 403–433. [DOI] [PubMed]
- Skálová, T., Dohnálek, J., Spiwok, V., Lipovová, P., Vondrácková, E., Petroková, H., Dusková, J., Strnad, H., Králová, B. & Hasek, J. (2005). J. Mol. Biol. 353, 282–294. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.