Table 2. Data collection and structure refinement.
Values in parentheses are for the highest resolution shell.
| Thaumatin (in loop, 100K) | Lysozyme (in device, room temperature) | |
|---|---|---|
| Data reduction | ||
| Wavelength () | 0.9537 | 0.9793 |
| Space group | P41212 | P43212 |
| Resolution () | 25.001.25 (1.271.25) | 20.001.65 (1.681.65) |
| Unit-cell parameters () | a = b = 57.91, c = 150.13 | a = b = 79.15, c = 38.02 |
| I/(I) | 31.0 (1.8) | 22.3 (2.0) |
| Completeness (%) | 99.7 (95.9) | 90.2 (94.3) |
| R merge † (%) | 5.3 (49.9) | 5.9 (47.4) |
| Multiplicity | 7.7 (3.6) | 2.9 (2.8) |
| Mosaicity () | 0.21 | 0.15 |
| Solvent content (%) | 48 | 31 |
| No. of frames | 360 | 35 |
| Oscillation per frame () | 0.3 | 1 |
| Refinement | ||
| Resolution () | 23.001.25 (1.281.25) | 19.201.65 (1.691.65) |
| R work ‡/R free § (%) | 15.4/16.9 | 17.2/22.2 |
| No. of protein residues/atoms | 206/1570 | 129/997 |
| No. of tartrate atoms | 10 [1 TAR¶] | 0 |
| No. of waters | 201 | 101 |
| Average B (2) | 12.00 | 21.67 |
| Protein only | 10.99 | 20.29 |
| Tartrate ion | 8.89 | |
| Solvent | 19.98 | 36.5 |
| R.m.s.d.†† | ||
| Bonds () | 0.012 | 0.015 |
| Angles () | 1.461 | 1.665 |
| PDB entry | 3qy5 | 3qy4 |
R
merge = 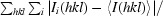
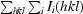 , where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and I(hkl) is its average.
, where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and I(hkl) is its average.
R = 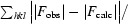
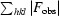 , where F
obs and F
calc are the observed and calculated structure factors, respectively.
, where F
obs and F
calc are the observed and calculated structure factors, respectively.
R free was calculated using 5% of the diffraction data, selected at random, which were excluded from refinement.
TAR refers to one tartrate ion.
Root-mean-square deviation.
