Fig. 9.
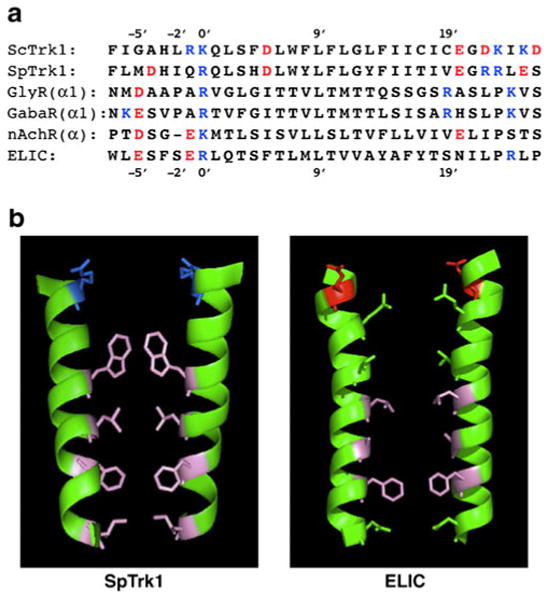
Comparison of yeast transmembrane segments M1D and pore-lining segments (M2, α2) of four ligand-gated ion channels (LGICs). a Primary sequences, in order top to bottom: S. cerevisiae; Schizosaccharomyces pombe, human glycine receptor α1 subunit, GABAA receptor α subunit (highly conserved sequence, identical in many species: human, mouse, rat, chicken, zebrafish, etc.); nicotinic acetylcholine receptor α subunit (also highly conserved: human, etc.); Erwinia chrysanthemi, recently crystallized bacterial homologue of nAchR [11, 36]. Customary numbering convention for LGICs, with 0′ lying at the internal face of the membrane and 20′ at the external face. b Ribbon models of paired transmembrane segments for SpTrk1 (left panel; the sequence directly modeled by Durell and Guy [23]) and for ELIC, spanning from −1′ at the inner surface of the membrane to 20′ at the outer surface. Stick figures depict only those side chains pointing into the channel. Blue basic amino acids, red acidic amino acids, pink hydrophobic amino acids, green uncharged polar amino acids. [The guanidinium side-chain on arginine at 0′ in ELIC projects away from the pore, and therefore is not shown. Bona fide LGIC channels are pentameric; the postulated structure for TRK-Cl− channels is tetrameric.] Drawing via PyMOL (DeLano Scientific, & Schrödinger LLC, Portland, OR, USA)
