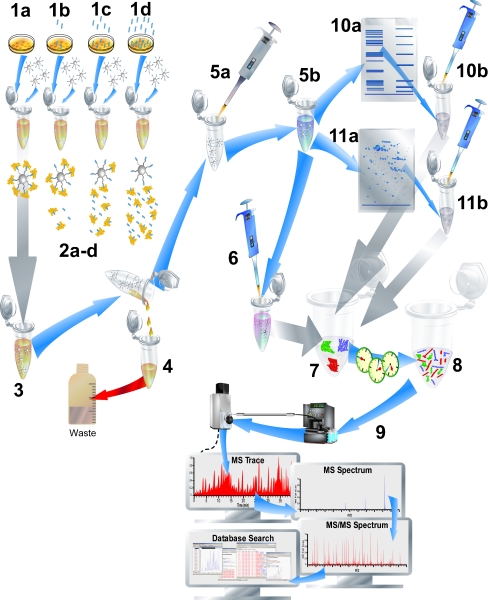
Fig. 4.
Chemical proteomics. The typical workflow is shown. The sample with different concentrations of the test ligand (four concentrations; 1a, 1b, 1c, and 1d) are subjected to lysates of cultured cells of interest and incubated with immobilized ligand affinity beads (2a-d). After incubation (3), the affinity beads with bound target proteins are removed from the incubation (4), washed, resuspended (5a), and finally a disruption step (5b) releases the bound target proteins from the beads. The crude protein mixtures are then prepared for 1D (10a) or potentially 2D (11a) gel electrophoresis followed by excision of the proteins (10b, 11b). The proteins can then be in-gel digested prior to analysis. Alternatively, proteins can be directly prepared for digestion (6), digested (7 and 8) and subsequently labeled with, e.g., isobaric tags for relative and absolute quantitation (iTRAQ). After iTRAQ labeling, samples are combined for straightforward eventual relative protein quantification from MS/MS spectra (see Fig. 5) obtained by LC-MS/MS analysis (9)