Abstract
Purpose
To evaluate Taylor dispersion analysis (TDA) as a novel method for determination of hydrodynamic radius of therapeutic peptides and proteins in non-stressed and stressed formulations and to compare it with dynamic light scattering (DLS).
Methods
The hydrodynamic radius of oxytocin, bovine serum albumin, various monoclonal antibodies (type IgG) and etanercept at concentrations between 0.05 and 50 mg/ml was determined by TDA and DLS. IgGs and etanercept were stressed (elevated temperatures) and analyzed by TDA, DLS and HP-SEC.
Results
TDA and DLS were comparable in sizing non-stressed peptides and proteins in a concentration range of about 0.5 to 50 mg/ml. TDA performed well even at lower concentrations, where DLS tends to provide theoretically high values of the Z-average radius. However, because of differences in the detection physics, DLS was more weighted towards the detection of aggregates in stressed formulations than TDA. Advantageously, TDA was also able to size the small peptide oxytocin, which was not feasible by DLS.
Conclusion
TDA allows the accurate determination of the hydrodynamic radius of peptides and proteins over a wide concentration range, with little interference from excipients present in the sample. It is marginally less sensitive than DLS in detecting size increase for stressed protein samples.
KEY WORDS: dynamic light scattering, hydrodynamic size, protein aggregation, protein characterization, Taylor dispersion analysis
INTRODUCTION
The size analysis of peptides and proteins in solution, and their associated degradation products (mainly aggregates), is a central aspect of analytical characterization during pharmaceutical development. Size information of peptides and proteins can be gained by separation-based methods, including analytical ultracentrifugation, size-exclusion chromatography (HP-SEC), asymmetrical flow field fractionation (FFF) and SDS-PAGE (1–3). The combination of HP-SEC and FFF with multi-angle laser light scattering detection (4,5) and analytical ultracentrifugation allows for the determination of both the molar mass and size without relying on molecular weight standards. A second important measurement principle is gaining size information by measuring the diffusion coefficient of the molecules in solution. The best known technique in the field of therapeutic proteins is dynamic light scattering (DLS), which determines the diffusion coefficient of molecules from the intensity fluctuations of scattered light on particles moving according to the Brownian motion (6). The hydrodynamic radius is then derived from the diffusion coefficient by the Stokes Einstein equation (2,6,7). DLS is a popular technique because it is user friendly, is widely available in pharmaceutical labs, and it enables sensitive aggregate detection. The fact that the intensity of the scattered light is proportional to the sixth power of the radius results in a bias of DLS to larger sizes (6). This can be an advantage, e.g., for the sensitive detection of small quantities of aggregates, or a disadvantage when aiming to size small peptides in general or monomeric protein in the presence of larger aggregates. Many new methods of analysis are being applied to solve the challenge of rapidly sizing biopharmaceutical systems using small quantities. For example, nanoparticle tracking analysis (NTA) is a novel imaging-based technique which measures the diffusion coefficient of individual particles by a microscope and camera (8). Although having several advantages over DLS, as described in detail by Filipe et al. (2010) (8), NTA is not suitable to size peptides or protein monomers, because they are too small for the size range of ca. 30 to 1,000 nm covered by NTA. Thus, the opportunity for new analytical tools to address such issues exists. In particular, methods that facilitate rapid analysis and screening and possess high accuracy and low sample consumption are highly desirable.
Taylor dispersion analysis (TDA) is a comparatively unutilized method to determine the diffusion coefficients and hydrodynamic radii of molecules. It is a fast, absolute method based on the dispersion of a solute plug through a uniform cylindrical tube under laminar Poiseuille flow. The method, sometimes termed Taylor-Aris dispersion, was first described by Taylor in 1953 (9) and developed further by Aris in 1956 (10). A plug of solute is injected into a moving solvent stream in an open tubular column which disperses by a combination of radial diffusion and cross-sectional velocity. So far, TDA has mainly been employed for nanoparticles (11), polymers (12–14) and small molecules (15,16). The reported use of TDA for protein characterization is rare. In 1994, Bello et al. sized the proteins ovalbumin and hemoglobin, but the emphasis of their work was to report the major advance of the use of small bore glass capillaries, radii ≤ 50 μm, which enabled the reduction of analysis time and sample consumption (17). More recently published, another rare example of a peer-reviewed article involving a protein, is a study on the interaction of the drug propranolol with serum proteins (18). Applications of TDA for the characterization of therapeutic proteins in complex formulations with typically used excipients have so far not been described in the literature.
In this paper, we present our evaluation of TDA for the determination of the hydrodynamic radius of therapeutic peptides and proteins in non-stressed and stressed formulations compared to DLS. The newly commercially available instrument for protein sizing (TDA200) utilizes pixilated UV area imaging to improve the quality of data collection, allowing TDA the potential to routinely measure the hydrodynamic radius of therapeutic proteins and peptides. The detector monitors broadening of a band of a therapeutic protein or small molecule solution injected into a stream of buffer solution and driven through a fused-silica capillary using a capillary electrophoresis type autosampler. The band is imaged at two windows, the first on entry to and the second on exit from a loop in the capillary. Band broadening due to Taylor dispersion is calculated from absorbance versus time data using the peak centre times at the first and second window, t1 and t2, respectively, and the corresponding standard deviations, τ1 and τ2 (band broadening), using Eq. 1:
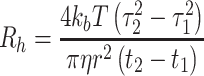 |
1 |
where R h is hydrodynamic radius, k b is the Boltzman constant, T is the temperature, η is the viscosity, and r is the radius of capillary.
Given the interest in the pathways to protein aggregation and analysis thereof (19), the need for analytical methods for aggregates and subvisible particles and for evaluating their performance is high (20). Our comparative evaluation is timely since new analytical tools suggested to be used for protein aggregation analysis still lack comparison to more established methods (19). Another recent commentary strongly supports the need for further work to assess the performance and limitations of methods to assess protein aggregates (21). The authors suggested that potential inaccurate quantitation and sizing of protein aggregates by size exclusion chromatography necessitate an essential need to use orthogonal analytical methods to assure protein product quality.
Within the context of methods to assess protein aggregation, the aim of our study was to evaluate the performance of Taylor dispersion analysis (TDA) as a novel method to determine the hydrodynamic radius of therapeutic proteins and peptides and their aggregates. We report the most extensive evaluation so far of TDA pertinent to the area of therapeutic proteins. Notably, oxytocin, bovine serum albumin, several monoclonal antibodies and etanercept are used as models, including stressed and un-stressed formulated products. The results obtained by TDA were compared to those of dynamic light scattering (DLS), which has been so far the standard technique for those applications in the pharmaceutical field. The influence of protein concentration, the presence of excipients, e.g. sugars, and protein aggregation on the measurement results were factors to be investigated using not only model proteins, but also formulated commercial proteins. Capabilities, advantages and limitations of Taylor dispersion analysis compared to DLS were identified and are discussed within the manuscript. Of particular interest was the probing of the relative ability of TDA to indicate formulated product change after thermal stress.
MATERIALS AND METHODS
Materials
The commercial protein products Enbrel®50 mg, Humira®40 mg and MabThera®100 mg were obtained from local hospitals (Amsterdam and Leiden, The Netherlands). A humanized IgG type 1 was used as a model protein (molar mass 150 kDa, isoelectric point ~8–9). Oxytocin and BSA were purchased from Sigma (Sigma Aldrich, Zwijndrecht, The Netherlands). All other substances used for preparation of buffers and mobile phase were standard analytical grades.
Enbrel®50 mg (50 mg/ml etanercept, 10 mg/ml sucrose, 5.8 mg/ml NaCl, 5.3 mg/ml arginine*HCl and 3.9 mg/ml Na2HPO4*2H20 pH 6.3) and Humira®40 mg (50 mg/ml adalimumab, 2 mg/ml NaCl, 1.52 mg/ml Na2HPO4*2H20, 0.3 mg/ml sodium citrate, 1.3 mg/ml citric acid monohydrate, 12.0 mg/ml mannitol and 1.0 mg/ml polysorbate 80 pH 5.2) were analyzed at different concentrations between 50 and 0.05 mg/ml. The formulations were diluted either with only buffer (3.9 mg/ml Na2HPO4*2H20 pH 6.3 for Enbrel®50 mg/ml and 1.52 mg/ml Na2HPO4*2H20, 0.3 mg/ml sodium citrate, 1.3 mg/ml citric acid monohydrate, pH 5.2 for Humira®40 mg) or the placebo buffer including all excipients. Mabthera®100 mg (10 mg/ml rituximab, 9 mg/ml NaCl, 7.3 mg/ml sodium citrate dehydrate and 0.7 mg/ml polysorbate 80, pH 6.5) was used at a concentration of 10 mg/ml and the humanized IgG1 (1.0 mg/ml IgG, 100 mM phosphate, pH 7.2) at 1 mg/ml. Oxytocin (50 mM phosphate, pH 4.5) was used in concentrations between 40 and 0.05 mg/ml and BSA (PBS, pH 7.2) between 50 and 0.05 mg/ml.
To induce aggregation, the commercial preparations and the humanized IgG1 were subjected to 10 min thermal stress using 1.5-ml reaction caps and a thermomixer (Eppendorf, Germany). Enbrel®50 mg was stressed at 65 and 70°C, Humira®40 mg and Mabthera®100 mg at 60 and 65°C and the model IgG at 75 and 80°C.
Taylor Dispersion Analysis
TDA was performed on a TDA200 HT nano-sizing sytem (Paraytec Ltd., York, UK). Samples (56 nl) were injected into fused silica capillary under a continuous flow of buffer (2 mm/s) using a CE injection system (PrinCE, Prince technologies B.V., Netherlands). The corresponding formulation buffer was used as running buffer. The detector head is placed inside the CE at a controlled temperature of 25°C. The total capillary length, with ID:OD dimensions of 75:360 μm, was 143.3 cm with length to first window being 46 cm and length between windows 48.5 cm. The capillary was cleaned between samples using a sodium hydroxide wash (1 M). UV detection wavelength used was 214 nm.
Dynamic Light Scattering
Dynamic light scattering (DLS) was performed on a Zetasizer Nano (Malvern, Herrenberg Germany). Five hundred μl of the formulations were analyzed in plastic cuvettes at 25°C using the automatic mode for identifying the best number of subruns and measurement time (n = 3). The z-average radius (zaverage) and polydispersity index (PDI) were calculated from the correlation function using the Dispersion Technology Software version 6.01 (Malvern, Herrenberg, Germany).
Size Exclusion Chromatography (HP-SEC)
The non-stressed and stressed samples of Enbrel®50 mg, Humira®40 mg, Mabthera®100 mg, and IgG were analyzed by HP-SEC, using a TSKgel4000SWXL column (Tosoh Biosep, Stuttgart, Germany) on an isocratic HPLC system with a Waters 515 pump, a Waters 717 plus autosampler, a Waters 474 fluorescence detector (Waters, Milford Massachusetts, USA) and a Shimadzu SPD UV/Vis detector (Shimadzu, Tokyo, Japan) at a flow rate of 0.5 ml/min. For the standard IgG formulations of 1.0 mg/ml, 50 μl sample was injected. The higher concentrated products were diluted to a concentration of 5 mg/ml using the particular formulation buffer, and 10 μl were injected. The mobile phase was composed of 50 mM sodium phosphate, 150 mM arginine and 0.025% NaN3, pH 6.5. To quantify aggregation in the IgG formulations, UV absorption at 280 nm was used.
Statistical Analysis
Statistical analysis was performed by a one-way ANOVA with a post-test (Tukey) using α = 0.05 (95% confidence interval) using GraphPad Prism (Version 5.02). Within the statistical analysis, the data (radii from TDA and DLS) for the stressed formulations were compared to the data of the non-stressed formulation as a control.
RESULTS AND DISCUSSION
Analysis of Peptides and Proteins Varying in Size and Concentration
No previous work has explored the use of TDA in its new format to determine the hydrodynamic radius of an extensive range of proteins and peptides differing in size (Table I) over a wide concentration range and in complex formulations containing various excipients. Oxytocin, BSA, etanercept and adalimumab formulated at concentrations between 0.05 and 50 mg/ml were analyzed by TDA and DLS, with the resulting average hydrodynamic radius rh (TDA), as well as the z-average radius rzave (DLS) and average main peak radius rmain peak (DLS) shown in Fig. 1. In comparing the relative strengths and weaknesses of the two methods, it is clear that TDA was able to provide consistent hydrodynamic radii for the smaller sized BSA and oxytocin at their lowest concentrations of 0.05 mg/ml. Both DLS and TDA were successful in sizing the larger sized antibody adalimumab at a low concentration of 0.05 mg/ml. For etanercept under the same conditions, neither technique was ideal for sizing purposes.
Table 1.
Peptides and Proteins Included into the Study
| Molar mass [kDa] | Literature values hydrodynamic radius [nm] | |
|---|---|---|
| Oxytocin | 1.007 | ~0.8 nm (30)a |
| BSA | 66.8 | 3.3–4.3 nm (23) |
| 3.7 nm (31) | ||
| Monoclonal antibodies: model IgG1, adalimumab, rituximab | ~150 | 5–6 nm (24–27) |
| Etanercept | ~150 | 7.1 nm (22) |
aradius estimated by Stokes-Einstein from diffusion coefficient of D = 4.34*10−6 cm2 s−1 as described in (30)
Fig. 1.
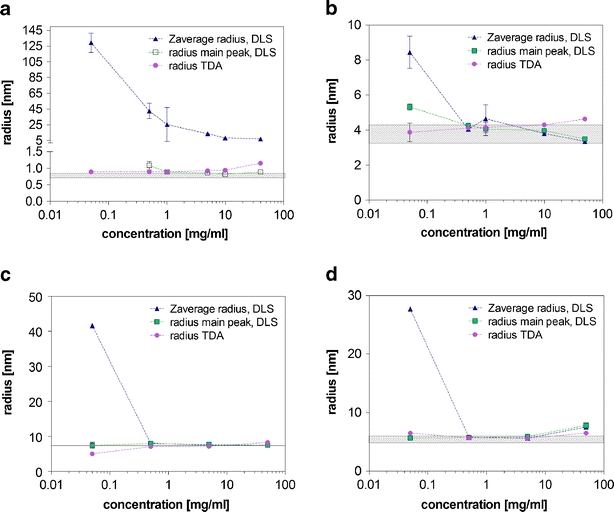
Radius from DLS (Zaverage radius, radius main peak) and Taylor dispersion analysis for oxytocin (a), BSA (b), etanercept (c) and adalimumab (d). The theoretical hydrodynamic radii (compare Table 1) are shown as light grey areas. The results are shown as average ± standard deviation of n = 3 (DLS) and n = 5 (TDA) individual measurements.
Considering each of the systems in more detail, for oxytocin the rh ranged from 0.9 to 1.1 nm when measured by TDA over all concentrations (Fig. 1a), which is slightly higher than the estimated hydrodynamic radius of about 0.8 nm (22). DLS proved to be less suitable for the analysis of oxytocin. Because of the small size of oxytocin and particularly at low concentrations, DLS analysis was influenced by the non-protein material in the formulation, e.g., small amounts of dust. Therefore, too high rzave values were measured (6.9–130 nm), in particular at the lower peptide concentrations. However, within the size distribution by intensity plots from DLS (data not shown), a peak at about 0.9 to 1 nm representing oxytocin could be identified for the concentration range of 1 to 40 mg/ml.
In the concentration range from 0.5 to 10 mg/ml, TDA and DLS resulted in similar results for the hydrodynamic radius of BSA of 3.8 to 4.3 nm (Fig. 1b), which is in good agreement with the theoretical size of 3.3 to 4.3 nm (23). At 50 mg/ml, TDA resulted in a higher rh of 4.6 nm, whereas rzave and rmain peak in DLS were found at 3.4 nm. At the lowest concentration of 0.05 mg/ml, rzave and rmain peak in DLS produced inconsistently higher radii values than theory (8.4 nm and 5.3 nm).
For etanercept, a hydrodynamic radius between 7.0 and 8.3 nm was measured by TDA and DLS in the concentration range from 0.5 to 50 mg/ml (Fig. 1c), which is in agreement with the theoretical value of 7.1 nm (22). For the formulation of etanercept at 0.05 mg/ml, TDA obtained a value rh of 4.9 nm, much lower than theoretically expected. For this sample and conditions, it would appear the TDA was operating at the limit of its detection. The rzave (DLS) was unexpectedly high (41.6 nm) for 0.05 mg/ml etanercept. In part, a possible explanation may be attributed to the influence of sucrose and other excipients at low protein concentration (discussed further later).
A hydrodynamic radius of about 5.6–5.9 nm was obtained by TDA and DLS for the concentration range from 0.5 to 5 mg/ml for adalimumab (Fig. 1d), which is the typical size for monomeric IgG (24–27). The slightly larger radius (7.5–8.3 nm) for the concentrated 50 mg/ml sample can be explained by reversible self-association, which is often observed for monoclonal antibody formulations at high concentrations (28). Again, the rzave determined for 0.05 mg/ml adalimumab was too high (27.7 nm), whereas rh (TDA) and rmain peak (DLS) were still accurate.
In the concentration range from 0.5 to 50 mg/ml, TDA and DLS were comparable to correctly determine the size of BSA, etanercept and adalimumab in the tested formulations. However, for the lowest concentrations of 0.05 mg/ml, TDA proved to be superior over DLS. The same behavior was identified for sizing of the small peptide oxytocin. When measuring formulations of low oxytocin concentrations, especially the rzave values in DLS were far too high. TDA was capable of correctly determining the correct size of oxytocin and the proteins at low concentrations.
This becomes also obvious when plotting the DLS results (rzave or rmain peak) against the rh (TDA), as shown in Fig. 2. For this comparison of TDA with DLS, additional measurements of rituximab, a model IgG, as well as etanercept and adalimumab diluted in buffer, were included. With the exception of the small peptide oxytocin findings and practically all formulations at 0.05 mg/ml, rzave (DLS) and rh (TDA) offered comparable results (Fig. 2a,b). The rmain peak (DLS) and rh (TDA) were in good agreement for all analyzed conditions (Fig. 2c).
Fig. 2.
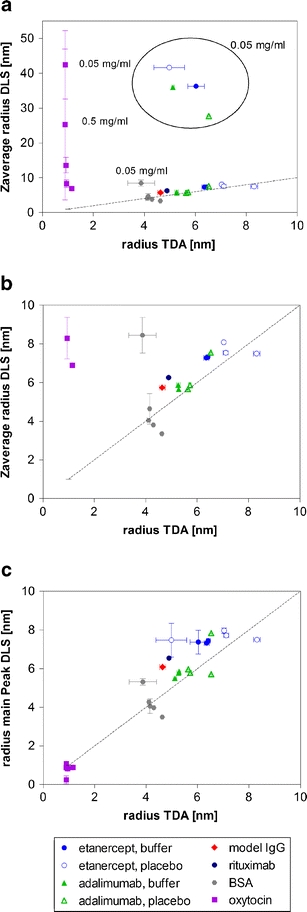
Comparison of radius from TDA with Zaverage radius from DLS (a, zoom as b), and the radius of the main peak from DLS (c) for the different peptides and proteins measured at concentrations between 0.05 and 50 mg/ml. The results are shown as average ± standard deviation of n = 3 (DLS) and n = 5 (TDA) individual measurements.
Influence of Excipients on the Analysis
The influence of excipients on protein characterization is an important issue, in particular as most therapeutic proteins are applied in complex formulations. DLS evaluates the scattering of not just the solute of interest in solution. As the above results confirm, with decrease in concentration and decrease in size of the substance of interest, DLS data can become more and more dominated by the scattering interference from excipients present in the formulations. Sugars and surfactants are often used as stabilizing excipients in therapeutic protein formulations. From previous studies for sugar solutions, e.g. sucrose, DLS typically measures two peaks corresponding to radii of about 0.5 nm and 100–200 nm (29). The reason for the size of 100–200 nm is not yet fully understood. To elucidate if TDA is also influenced by sugars, the etanercept formulation, which contains 1% (w/v) sucrose, was analyzed at etanercept concentrations between 0.05 and 50 mg/ml (Fig. 3a). For the protein-free placebo, DLS resulted in an rzave of 288 nm, a main peak at about 200 nm and a second peak <1 nm. In TDA, only a rh of 0.5 nm was measured for the sucrose-containing placebo, whereas the larger size of 200 nm was not detected. Sucrose also disturbed the DLS measurements at the low etanercept concentration of 0.05 mg/ml, where a too high rzave was measured. At higher protein concentrations above 0.5 mg/ml, the protein signal dominates, and the presence of sucrose no longer affects the rzave.
Fig. 3.
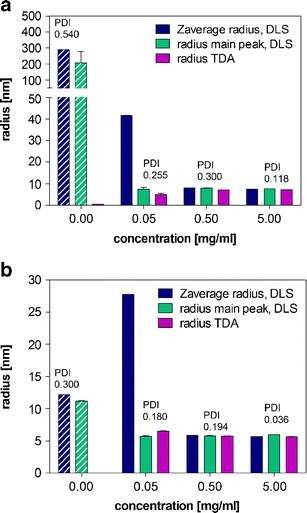
Radius (Zaverage radius, radius main peak) and polydispersity index (PDI) from DLS and Taylor dispersion analysis for etanercept (a) and adalimumab (b) diluted with formulation placebo. The results are shown as average ± standard deviation of n = 3 (DLS) and n = 5 (TDA) individual measurements.
Surfactants are a second class of excipients that can disturb DLS measurements, when used above the critical micelle concentration. This is also the case for the adalimumab placebo, which contains 0.1% polysorbate 80. Here, a rzave of about 12 nm was determined by DLS, corresponding to the polysorbate micelles (Fig. 3b). No rh could be resolved for the formulation placebo by TDA, as the formulation buffer was also used as a running buffer for the system.
The experiments showed that excipients like sugars and surfactants can disturb the size analysis of proteins by DLS, particularly for formulations with low protein concentrations. TDA has more selectivity, on the other hand, and its results were not affected to the same extent by the presence of excipients.
Analysis of Stressed Protein Formulations
TDA and DLS have the advantage that they can be used in batch mode over a range of concentration without the need for prior separation of components and without the need for internal standards. The ability to combine stress testing of protein formulations and rapidly detect any aggregation tendency is an important part of the biopharmaceutical development process. In this section of the work, we wished to see if TDA was able to detect change in the average size of stressed protein samples in a similar manner to DLS. Both techniques were benchmarked using HP-SEC to prove the formation of soluble aggregates. One advantage of TDA over DLS would be its lower injection sample size (previously optimized at 60 nl). TDA and DLS were compared for their ability to detect aggregation in heat-stressed formulations of etanercept, adalimumab, rituximab and a model IgG (Fig. 4), as well as in etanercept samples during a one-week accelerated stability study at 50°C (Fig. 5).
Fig. 4.
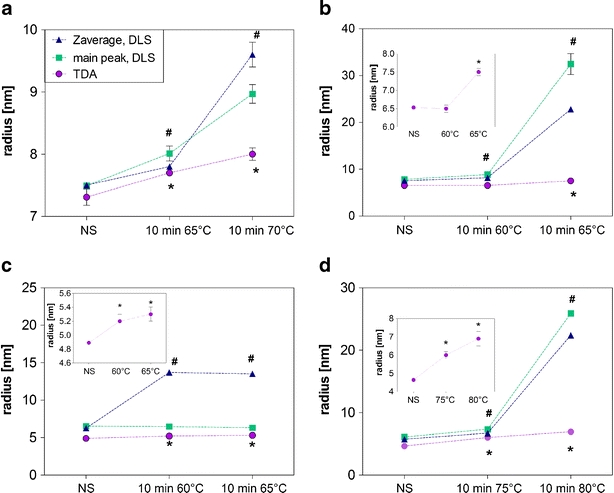
Results for DLS (rZave, rmain peak) and TDA (rh) for non-stressed and aggregated formulations of etanercept (a), adalimumab (b), rituximab (c) and the model IgG (d). *marks rh from TDA, # Zaverage values from DLS, which are significantly higher for the stressed than for the non-stressed formulation (p < 0.05). The results are shown as average ± standard deviation of n = 3 (DLS) and n = 5 (TDA) individual measurements.
Fig. 5.
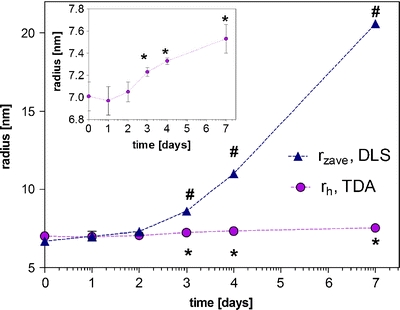
Results for DLS (rZave) and TDA (rh) for etanercept subjected to a 1-week stability study at 50°C. *marks rh from TDA, # Zaverage values from DLS, which are significantly higher for the stressed than for the non-stressed formulation (p < 0.05). The results are shown as average ± standard deviation of n = 3 (DLS) and n = 5 (TDA) individual measurements.
Heat stress induced the formation of soluble and non-soluble aggregates within the formulations, as shown by HP-SEC (Table II). In general, rzave and rmain peak (DLS) increased to a greater extent than rh (TDA) for the aggregated formulations (Fig. 4). This is mainly obvious for adalimumab stressed at 65°C (Fig. 4b) and the model IgG stressed at 80°C (Fig. 4d), which contained about 60% soluble and non-soluble aggregates as measure by HP-SEC (Table II). The rzave (DLS) increased from 5.7 nm (adalimumab) and 7.5 nm (IgG) to about 22 nm, whereas the rh (TDA) changed from 4.6 to 6.9 nm (adalimumab) and from 6.5 to 7.5 nm (IgG) after stressing. For formulations that mainly contained small soluble aggregates, the difference between rzave (DLS) and rh (TDA) was less pronounced.
Table II.
Size Analysis of Protein Samples by HP-SEC Quantified from UV Detection at 280 nm
| Monomer [%] | Dimer [%] | HMW [%] | Fragments [%] | Recovery [%] | Soluble aggregates + non-recovered fraction [%] | |
|---|---|---|---|---|---|---|
| Etanercept, NS | 93.0 | 1.3 | 0.0 | 5.6 | 100.0 | 7.0 |
| Etanerecept, 10 min 65°C | 88.9 | 3.5 | 2.0 | 5.6 | 96.6 | 14.1 |
| Etanerecept, 10 min 70°C | 81.3 | 6.0 | 7.4 | 5.3 | 95.8 | 22.1 |
| Adalimumab, NS | 99.3 | 0.4 | 0.0 | 0.3 | 100.0 | 0.7 |
| Adalimumab, 10 min 60°C | 96.8 | 1.0 | 1.8 | 0.3 | 101.6 | 1.7 |
| Adalimumab, 10 min 65°C | 58.3 | 1.8 | 39.5 | 0.3 | 69.6 | 59.4 |
| IgG, NS | 99.1 | 0.0 | 0.0 | 0.9 | 100.0 | 0.9 |
| IgG, 10 min 75°C | 95.4 | 3.0 | 0.0 | 1.6 | 94.8 | 9.6 |
| IgG, 10 min 80°C | 45.0 | 8.0 | 44.3 | 2.7 | 87.2 | 60.8 |
| Rituximab, NS | 99.4 | 0.6 | 0.0 | 0.0 | 100.0 | 0.6 |
| Rituximab, 10 min 60°C | 98.9 | 0.7 | 0.4 | 0.0 | 100.5 | 0.6 |
| Rituximab, 10 min 65°C | 97.3 | 1.0 | 1.7 | 0.0 | 98.5 | 4.2 |
Whilst the absolute values of the average radii of the samples reported by TDA and DLS may have differed markedly on occasion, it is noteworthy that TDA was able to identify that all four tested proteins showed significant increase in size after undergoing the more aggressive stress test. After exposure of the four protein formulations to the less aggressive stress conditions, TDA results showed significant increase in size for three of the four formulations, whereas DLS showed all four to be significant. On inspection of the HP-SEC data, this equates to TDA being able to statistically significantly differentiate between a sample of rituximab containing 99.4% monomer and one containing 98.9% monomer but not able to discriminate between a sample of adalimumab containing 99.3% monomer and one containing 96.8%. Possible contributory factors for this difference may include the fact that the samples were diluted for the HP-SEC analysis, whereas the TDA and DLS samples were run without dilution.
Stress studies can form part of the drug development process but would generally use lower temperatures than that employed above. A comparison of the ability of TDA and DLS to identify size changes after a longer isothermal temperature hold was undertaken. During a one-week stability study of etanercept performed at 50°C, a comparable observation to those obtained in the previous paragraph was made: the rzave (DLS) increased to a higher level, i.e. from 6.7 nm to 20.6 nm after 7 days at 50°C, than the rh (TDA), which increased from 7.0 to 7.5 nm. However, for both DLS and TDA the increases in rzave (DLS) and rh (TDA) were found to be significant at day 3 of the stability study.
The differences between DLS and TDA in their apparent sensitivity to detect aggregation can be explained by the different underlying measurement principles of the two techniques. DLS is biased to larger sizes, because the intensity of the scattered light is proportional to the sixth power of the radius (6). In particular, larger aggregates will contribute more to the overall scattering intensity, and the rzave is shifted to larger sizes. In TDA, the diffusion coefficient is derived from the band broadening measured by UV absorbance. For a molar concentration-based detector system, it can be assumed that monomeric and aggregated protein molecules are detected equally (neglecting the contribution of light scattering to the optical density at this point) and thus averaging of the hydrodynamic radii of mixtures is a number average (13). The inference is that TDA is more weighted and suited to studies where the focus is protein-protein interactions and DLS will more readily identify the larger sized aggregates that their scattering by intensity produces.
CONCLUSIONS
This work has explored the use of TDA in its new format linked to a pixilated area detector and compared it with DLS to determine the hydrodynamic radius of an extensive range of peptide and protein sizes in therapeutic formulations over a wide concentration range and after stress testing. Our findings suggest that DLS and TDA have some applications of overlap but that their different physical basis for the determination of size gives rise to different strengths for the sizing of biopharmaceuticals and their aggregates. For relatively pure unstressed protein formulations (BSA, etancercept and monoclonal antibodies), TDA and DLS resulted in comparable results for the hydrodynamic radius in the concentration range from 0.5 to 50 mg/ml. Lower protein concentrations (0.05 mg/ml), as well as the small peptide oxytocin, could only be sized correctly by TDA, whereas DLS resulted in much larger values for the radius than theoretically expected. Another advantage of TDA is that it can be selectively tuned to detect the protein at low concentration and obviate the strong contributory factors that some excipients can make to DLS results. When focusing on the detection of aggregation after stress testing, both DLS and TDA identified significant size change for three protein systems after stressing. TDA compared to DLS was less sensitive in a fourth case. An explanation lies in the fact that the underlying principle of DLS is weighted to the detection of larger particles, and as a consequence also explains the differences in the absolute values obtained for the two methods.
ACKNOWLEDGMENTS
This research was supported, in part, by the Dutch Technology Foundation STW, Applied Science Division of NWO and the Technology Program of the Ministry of Economic Affairs. The authors also acknowledge the EPSRC grant (DT/F002564/1) for funding as part of a TSB collaborative research project.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Andrea Hawe and Wendy L. Hulse share first author.
REFERENCES
- 1.Fraunhofer W, Winter G. The use of asymmetrical flow field-flow fractionation in pharmaceutics and biopharmaceutics. Eur J Pharm Biopharm. 2004;58:369–83. doi: 10.1016/j.ejpb.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 2.Philo JS. Is any measurement method optimal for all aggregate sizes and types? AAPS J. 2006;8:E564–71. doi: 10.1208/aapsj080365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Philo JS. A critical review of methods for size characterization of non-particulate protein aggregates. Curr Pharm Biotechnol. 2009;10:359–72. doi: 10.2174/138920109788488815. [DOI] [PubMed] [Google Scholar]
- 4.Kendrick BS, Kerwin BA, Chang BS, Philo JS. Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein-protein or protein-ligand association states. Anal Biochem. 2001;299:136–46. doi: 10.1006/abio.2001.5411. [DOI] [PubMed] [Google Scholar]
- 5.Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–66. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- 6.Demeester J, De Smedt S, Sanders N, Haustraete J. Light scattering. In: Jiskootand W, Crommelin DJA, editors. Methods for structural analysis of protein pharmaceuticals. Arlington: AAPS; 2005. [Google Scholar]
- 7.Nobbmann U, Connah M, Fish B, Varley P, Gee C, Mulot S, Chen J, Zhou L, Lu Y, Shen F, Yi J, Harding SE. Dynamic light scattering as a relative tool for assessing the molecular integrity and stability of monoclonal antibodies. Biotechnol Genet Eng Rev. 2007;24:117–28. doi: 10.1080/02648725.2007.10648095. [DOI] [PubMed] [Google Scholar]
- 8.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor GA. Dispersion of soluble matter in solvent flowing slowly through a tube. Proc R Soc Lond Ser A Math Phys Eng Sci. 1953;219:186–203. doi: 10.1098/rspa.1953.0139. [DOI] [Google Scholar]
- 10.Aris R. On the dispersion of a solute in a fluid flowing through a tube. Proc R Soc Lond Ser A Math Phys Eng Sci. 1956;235:67–77. doi: 10.1098/rspa.1956.0065. [DOI] [Google Scholar]
- 11.d’Orlye F, Varenne A, Gareil P. Determination of nanoparticle diffusion coefficients by Taylor dispersion analysis using a capillary electrophoresis instrument. J Chromatogr A. 2008;1204:226–32. doi: 10.1016/j.chroma.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Cottet H, Biron JP, Cipelletti L, Matmour R, Martin M. Determination of individual diffusion coefficients in evolving binary mixtures by taylor dispersion analysis: application to the monitoring of polymer reaction. Anal Chem. 2010;82:1793–802. doi: 10.1021/ac902397x. [DOI] [PubMed] [Google Scholar]
- 13.Cottet H, Martin M, Papillaud A, Souaid E, Collet H, Commeyras A. Determination of dendrigraft poly-L-lysine diffusion coefficients by taylor dispersion analysis. Biomacromolecules. 2007;8:3235–43. doi: 10.1021/bm070268j. [DOI] [PubMed] [Google Scholar]
- 14.Wernert V, Bouchet R, Denoyel R. Influence of molecule size on its transport properties through a porous medium. Anal Chem. 2010;82:2668–79. doi: 10.1021/ac902858b. [DOI] [PubMed] [Google Scholar]
- 15.Cottet H, Biron JP, Martin M. Taylor dispersion analysis of mixtures. Anal Chem. 2007;79:9066–73. doi: 10.1021/ac071018w. [DOI] [PubMed] [Google Scholar]
- 16.Sharma U, Gleason NJ, Carbeck JD. Diffusivity of solutes measured in glass capillaries using Taylor’s analysis of dispersion and a commercial CE instrument. Anal Chem. 2005;77:806–13. doi: 10.1021/ac048846z. [DOI] [PubMed] [Google Scholar]
- 17.Bello MS, Rezzonico R, Righetti PG. Use of taylor-aris dispersion for measurement of a solute diffusion coefficient in thin capillaries. Science. 1994;266:773–6. doi: 10.1126/science.266.5186.773. [DOI] [PubMed] [Google Scholar]
- 18.Ostergaard J, Jensen H. Simultaneous evaluation of ligand binding properties and protein size by electrophoresis and Taylor dispersion in capillaries. Anal Chem. 2009;81:8644–8. doi: 10.1021/ac901419x. [DOI] [PubMed] [Google Scholar]
- 19.Mahler HC, Friess W, Grauschopf U, Kiese S. Protein aggregation: pathways, induction factors and analysis. J Pharm Sci. 2009;98:2909–34. doi: 10.1002/jps.21566. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJA, Middaugh CR, Winter G, Fan YX, Kirshner S, Verthelyi D, Kozlowski S, Clouse KA, Swann PG, Rosenberg A, Cherney B. Overlooking subvisible particles in therapeutic protein products: gaps that may compromise product quality. J Pharm Sci. 2009;98:1201–5. doi: 10.1002/jps.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJ, Middaugh CR, Winter G. Potential inaccurate quantitation and sizing of protein aggregates by size exclusion chromatography: essential need to use orthogonal methods to assure the quality of therapeutic protein products. J Pharm Sci. 2010;99:2200–8. doi: 10.1002/jps.21989. [DOI] [PubMed] [Google Scholar]
- 22.Kohno T, Tam LTT, Stevens SR, Louie JS. Binding characteristics of tumor necrosis factor-Fc fusion proteins vs anti-tumar necrosis factor mAbs. J Investig Dermatol Symp Proc. 2007;2007:5–8. doi: 10.1038/sj.jidsymp.5650034. [DOI] [PubMed] [Google Scholar]
- 23.Jachimska B, Wasilewska M, Adamczyk Z. Characterization of globular protein solutions by dynamic light scattering, electrophoretic mobility, and viscosity measurements. Langmuir. 2008;24:6866–72. doi: 10.1021/la800548p. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, Graf O, Milovic N, Luan X, Bluemel M, Smolny M, Forrer K. Formulation development of antibodies using robotic system and high-throughput laboratory (HTL) J Pharm Sci. 2010;99:2279–94. doi: 10.1002/jps.22008. [DOI] [PubMed] [Google Scholar]
- 25.Bermudez O, Forciniti D. Aggregation and denaturation of antibodies: a capillary electrophoresis, dynamic light scattering, and aqueous two-phase partitioning study. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;807:17–24. doi: 10.1016/j.jchromb.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 26.Sukumar M, Doyle BL, Combs JL, Pekar AH. Opalescent appearance of an IgG1 antibody at high concentrations and its relationship to noncovalent association. Pharm Res. 2004;21:1087–93. doi: 10.1023/B:PHAM.0000032993.98705.73. [DOI] [PubMed] [Google Scholar]
- 27.Hawe A, Friess W, Sutter M, Jiskoot W. Online fluorescent dye detection method for the characterization of immunoglobulin G aggregation by size exclusion chromatography and asymmetrical flow field flow fractionation. Anal Biochem. 2008;378:115–22. doi: 10.1016/j.ab.2008.03.050. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Nguyen MD, Andya JD, Shire SJ. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J Pharm Sci. 2005;94:1928–40. doi: 10.1002/jps.20347. [DOI] [PubMed] [Google Scholar]
- 29.Kaszuba M, McKnight D, Connah MT, McNeil-Watson FK, Nobbmann U. Measuring sub nanometre sizes using dynamic light scattering. J Nanopart Res. 2008;10:823–9. doi: 10.1007/s11051-007-9317-4. [DOI] [Google Scholar]
- 30.Hosoya O, Chono S, Saso Y, Juni K, Morimoto K, Seki T. Determination of diffusion coefficients of peptides and prediction of permeability through a porous membrane. J Pharm Pharmacol. 2004;56:1501–7. doi: 10.1211/0022357044878. [DOI] [PubMed] [Google Scholar]
- 31.Brownsey GJ, Noel TR, Parker R, Ring SG. The glass transition behavior of the globular protein bovine serum albumin. Biophys J. 2003;85:3943–50. doi: 10.1016/S0006-3495(03)74808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


