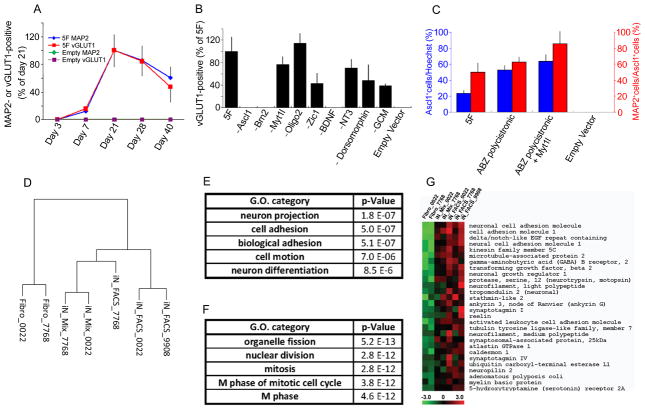
Figure 2. Further description of hiN cell conversion: essential factors and transcriptome analysis.
(A) Temporal profile of hiN cell conversion. MAP2- or vGLUT1-positive cells were quantified at indicated time points after transduction with conversion factor vectors (5F; indicated as blue or red line) or empty vector (Empty; green or purple line). The number of MAP2 (diamond) and vGLUT1 (square) -positive cells peaked at 21 days after 5F transduction, whereas such cells were not apparent with empty vector. N=3 at each time point; data are presented as mean ± SEM.
(B) Required factors in hiN cell conversion. Fibroblasts were transduced with the 5-factor (5F) cocktail as above, or with factor mixes lacking the indicated individual factors. Bar graphs indicate the number of vGLUT1-positive cells at 3 weeks after transduction, as a percent of 5F transduction. GCM, glial-conditioned media. N=3 per group.
(C) Fibroblasts were transduced with a polycistronic vector harboring Ascl1, Brn2, and Zic1 (ABZ-polycistronic) alone or in combination with a Myt1l vector. The percentage of Ascl1-positive cells per total cell number (Hoechst positive nuclei; blue bars) reflects the transduction efficiency. The percentage of MAP2-positive hiN cells of transduced Ascl1-positive cells (red bars) reflects the hiN cell conversion efficiency. N=3 per group.
(D) Dendrogram presenting the hierarchical clustering of gene expression array profiles as measured by Human Genome U133 Plus 2.0 Arrays (Affymetrix). Complete linkage hierarchical clustering analysis was performed using Pearson’s correlation metric. The dendrogram includes individual samples from FACS-sorted hiN cells (iN_FACS), unsorted hiN cell cultures (iN_Mix), or the original fibroblasts (Fibro). Samples are labeled as to the fibroblast of origin (see Table S1). hiN cell preparations clustered together, rather than with the originating fibroblast preparations.
(E and F) The five most significantly enriched gene ontology (GO) categories among the genes upregulated (E) or downregulated (F) in the context of hiN cell conversion are presented. Expression data were analyzed using a False Discovery Rate of less than 25% and a log-ratio threshold of >2. Nominal p-values are listed.
(G) Heat map specifying the genes and expression values within the GO category “Neuron projection” as in (E). Relative expression levels of individual genes (as labeled on rows) are presented from low (green) to high (red) as per the color chart bar at the bottom. Cell samples are labeled as per the (D). See also Figures S2 and S3 and Table S2.