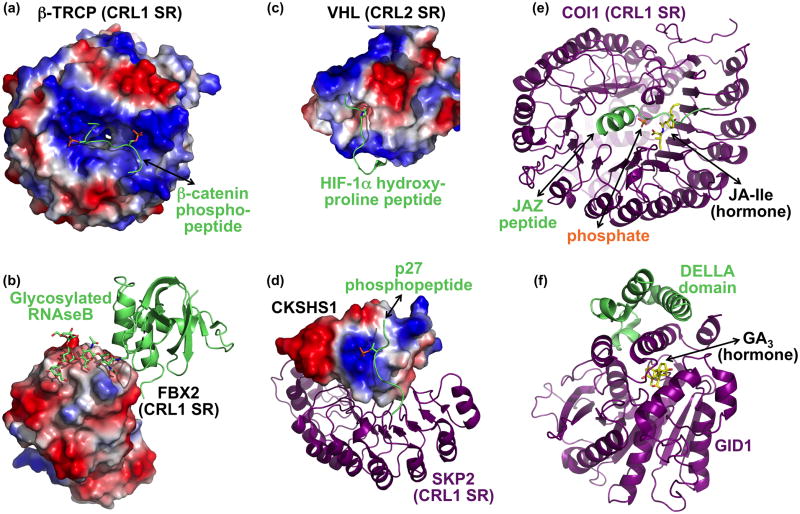
Figure 3. Selected examples of regulated substrate binding to CRLs.
(a) Two precisely positioned basic patches and a peptide binding groove in the CRL1 SR β-TRCP (surface showing electrostatic potential) recruit a doubly-phosphorylated β-catenin (lime, with phosphates as orange/red sticks) [10]. (b) Sugar recognition on a glycosylated substrate (lime) by the CRL1 SR FBX2 (electrostatic surface) [35]. The sugar moiety is depicted in sticks. (c) Recognition of HIF-1α hydroxyproline (peptide in lime, with hydroxyproline in sticks) by the CRL2 SR VHL (electrostatic surface) [32,33]. Only a portion of the VHL structure is shown to highlight substrate recognition. (d) The CRL1 SR Skp2 (purple) imports phosphopeptide recognition by binding to CKSHS1 (electrostatic surface), whose anion binding pocket recognizes the phosphate moiety on the substrate phospho-p27 (lime, with phosphate in orange/red sticks) [39]. (e) The plant hormone jasmonic acid-isoleucine (JA-Ile) functions in a multicomponent molecular glue mechanism to recruit substrates to the CRL1 SR COI-1 (purple). JA-Ile (yellow) mediates binding to the substrate JAZ degron peptide (lime), but the JAZ degron helix also clamps JA-Ile into the COI-1 binding site. A nearby phosphate most likely resembles part of inositol pentakisphosphate, which potentiates interaction between COI-1, JA-Ile, and a JAZ substrate. (f) Gibberellic acid (GA, yellow) uses an indirect allosteric mechanism to promote interactions between DELLA transcriptional repressors (lime) and a multiprotein CRL1 SR that includes GID1 (purple) [42]. GA serves as part of a hydrophobic core required for folding of the GID1 N-terminus. This portion of GID1, in turn, helps recruit DELLA substrates for ubiquitination.