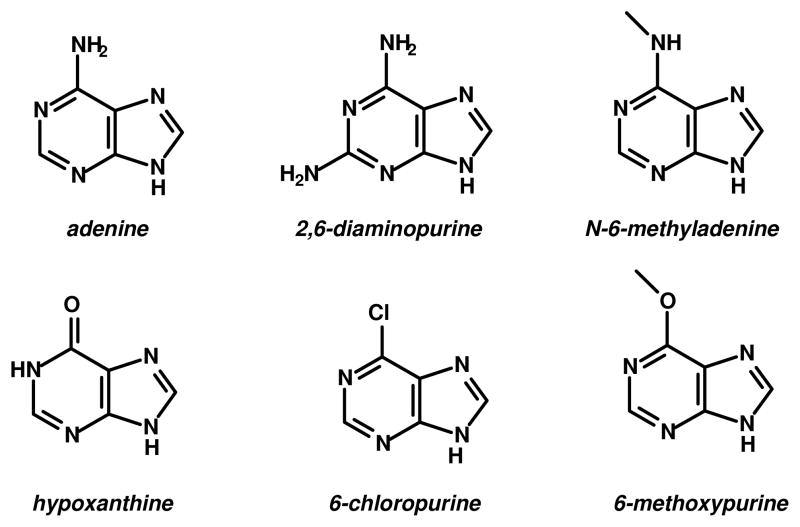
Abstract
Four proteins from NCBI cog1816, previously annotated as adenosine deaminases, have been subjected to structural and functional characterization. Pa0148 (Pseudomonas aeruginosa PAO1), AAur1117 (Arthrobacter aurescens TC1), Sgx9403e, and Sgx9403g, have been purified and their substrate profiles determined. Adenosine is not a substrate for any of these enzymes. All of these proteins will deaminate adenine to produce hypoxanthine with values of kcat/Km that exceed 105 M−1s−1. These enzymes will also accept 6-chloropurine, 6-methoxypurine, N-6-methyladenine, and 2,6-diaminopurine as alternate substrates. X-ray structures of Pa0148 and AAur1117 have been determined and reveal nearly identical distorted (β/α)8-barrels with a single zinc ion that is characteristic of members of the amidohydrolase superfamily. Structures of Pa0148 with adenine, 6-chloropurine and hypoxanthine were also determined thereby permitting identification of the residues responsible for coordinating the substrate and product.
According to NCBI, the genomes of more than 1,500 bacteria have been completely sequenced. Due to the large and rapidly increasing volume of DNA sequence data, functional annotations based primarily on protein similarity scores to enzymes of known function are now routinely employed (1). Unfortunately, the reliability of these functional annotations is directly dependent on the stringency of a given function for relatively small differences in amino acid sequence. Misannotations can occur from the utilization of similarity scores below reliable cutoff thresholds and from perturbations in catalytic function with relatively few changes in amino acid sequence (2). These problems are further amplified when the functional boundaries between orthologs and paralogs are difficult to discern based on sequence data alone (3). Misannotations are subsequently propagated to other databases and the number of proteins with incorrectly identified functions has increased (2). The considerable number of proteins of unknown or uncertain function suggests that a significant segment of the metabolic landscape remains undiscovered.
Adenosine deaminase is a member of the amidohydrolase superfamily (AHS) of enzymes. This superfamily was first identified by Holm and Sander based on the similarities in the three-dimensional structures of phosphotriesterase, adenosine deaminase and urease (3). Proteins in the AHS have a distorted (β/α)8-barrel fold with conserved metal binding residues at the C-terminal ends of β-strands 1, 4, 5, 6 and 8. These enzymes have either a mononuclear or binuclear metal center which activates a hydrolytic water for nucleophilic attack on amino acids, sugars, nucleic acids and organophosphate esters (4). Within the AHS there are 24 clusters of orthologous groups (COG) as defined by NCBI. Three of these clusters of enzymes are known to catalyze deamination reactions: cog1001, cog0402 and cog1816. The prototypical adenine deaminase (ADE) and N-6-methyladenine deaminase (6-MAD) are members of cog1001 (5). Enzymes that deaminate guanine, cytosine, S-adenosylhomocysteine, thiomethyl adenosine, N-formimino-L-glutamate, and 8-oxoguanine are found in cog0402 (6). According to NCBI, all bacterial proteins in cog1816 catalyze the deamination of adenosine.
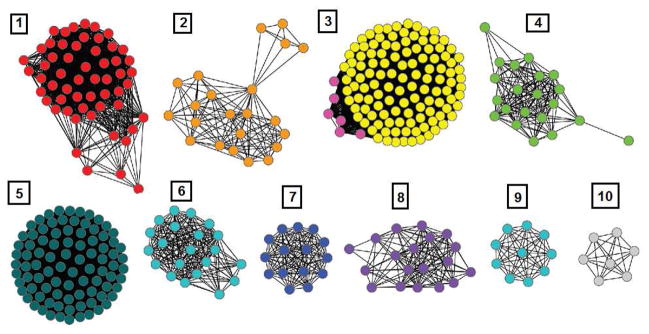
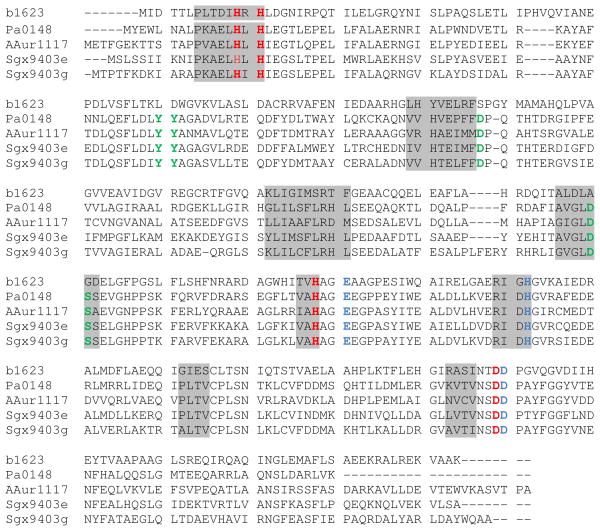
The structure and mechanism of action of adenosine deaminases from several organisms have been studied (7–19). A sequence similarity network for cog1816 is illustrated in Figure 1 (20). Enzymes experimentally verified as adenosine deaminase (ADD) are found in Group 5, which contains the adenosine deaminase from E. coli K-12 (locus tag: b1623). Essential residues for adenosine deaminase activity include an HxHxD motif following β-strand 1, an aspartate following β-strand 3, a glycine following β-strand 4, an HxxE motif following β-strand 5, a histidine following β-strand 6, and a pair of aspartates following β-strand 8 (8, 17, 19, 21, 22). These residues occur in every protein of Group 5. Despite an overall amino acid sequence identity of ~30% to the E. coli adenosine deaminase, proteins of Group 3 are missing several residues that have been implicated in substrate binding. These residues include aspartate residues following β-strands 1 and 3, and the glycine residue following β-strand 4. Based on sequence alignments between Group 3 and 5 enzymes, we anticipated that some members of cog1816 do not catalyze the deamination of adenosine.
Figure 1.
Sequence similarity network created using Cytoscape (www.cytoscape.org) of cog1816 from the amidohydrolase superfamily. Each node in the network represents a single sequence, and each edge (depicted as lines) represents the pairwise connection between two sequences at a BLAST E-value of better than 1 × 10−70. Lengths of edges are not significant, except for tightly clustered groups, which are more closely related than sequences with only a few connections. Group 5 contains the adenosine deaminase from E. coli. In Group 3, the sequences depicted in yellow contain all of the essential residues for the deamination of adenine identified in this investigation.
The initial target for this investigation was the functional annotation of Pa0148 from Pseudomonas aeruginosa PAO1. This enzyme and three other enzymes are in Group 3 of cog1816. The substrate profiles for all four enzymes have been determined and the three-dimensional structures of Pa0148 and Aaur1117 from Arthrobacter aurescens TC1 have been determined with substrate and product ligands bound in the active site. These enzymes do not catalyze the deamination of adenosine but they do catalyze the deamination of adenine to hypoxanthine.
MATERIALS AND METHODS
Materials
All chemicals were purchased from Sigma-Aldrich, unless otherwise stated. N-6-methyladenine was purchased from Spectrum and 6-methoxypurine was obtained from Tokyo Chemical Industry Co. Zeatin was acquired from MP Biomedicals and isoguanine was bought from Santa Crux Biotechnology. 2′-Deoxy-N-6-methyladenosine was procured from Carbosynth.
Cloning and Purification of Pa0148, Sgx9403e and Sgx9403g
The gene for Pa0148 (gi| 15595346; Sgx9608a) was obtained from the genomic DNA of P. aeruginosa PAO1 (ATCC 47085D), and the genes for Sgx9403e and Sgx9403g were chemically synthesized by back-translation and codon optimized for E. coli expression (Codon Devices, Inc.). The latter two genes were first identified from the Sargasso Sea environmental sequencing effort through the Global Ocean Sampling expedition and the original GI numbers (44392365 and 44590840 for Sgx9403e and Sgx9403g, respectively) are now obsolete. The sequence currently in Genbank that is most similar to Sgx9403e is gi|239993551, which belongs to AmacA2_3576 from Alteromonas macleodii ATCC 27126. With the exception of the first 16 residues of Sgx9403e that are not present in AmacA2_3576, the two proteins are 99% identical. The most similar sequence in Genbank to Sgx9403g is gi| 78065603, which belongs to Bcep18194_A4131 from Burkholderia sp. 383. Bcep18194_A4131 and Sgx9403g share a 99% sequence identity. The synthesized genes were cloned into a custom TOPO-isomerase vector, pSGX3(BC), supplied by Invitrogen. Forward primers for Pa0148, Sgx9403e, and Sgx9403g were TACGAATGGCTCAACGCCTTG, AGCCTGAGCAGCATTATCAAAAATAT, and ACCCCGACCTTTAAAGACAAG, respectively, and reverse primers were CCTTCACCAGGCGCGCATCCA, CTGCTGAGAGTACTTTTTCAACC, and CTGCAGCTTGCCAATACGCATC, respectively. The clones encode Met-Ser-Leu followed by the PCR product and end with Glu-Gly-His6. Miniprep DNA was transformed into BL21(DE3)-Codon+RIL expression cells (Stratagene), expressed, and made into a 30% glycerol stock for large-scale fermentation.
The expression clones were cultured using High Yield selenomethionine (SeMet) media (Orion Enterprises, Inc., Northbrook, IL). Overnight cultures (50 mL) in 250 mL baffled flasks were cultivated at 37 °C from a frozen glycerol stock for 16 hours, then transferred to 2 L baffled shake flasks containing 1 L HY-SeMet media (100 μg/mL kanamycin and 30 μg/mL chloramphenicol) and grown to OD600 ~ 1.0. SeMet was then added for labeling at 90 mg/L, followed by IPTG added to 0.4 mM final concentration. Cells were further grown at 22 °C for 18 hours, then harvested using standard centrifugation for 10 minutes at 6000 rpm and frozen at −80 °C.
Cells were lysed in 20 mM Tris, pH 8.0, 0.5 M NaCl, 25 mM imidazole, and 0.1% Tween20 by sonication. The cellular debris was removed by centrifugation for 30 minutes (39,800g). The supernatant was collected and incubated with 10 mL of a 50% slurry of Ni-NTA agarose (Qiagen) for 30 minutes with gentle stirring. The sample was then poured into a drip column and washed with 50 mL of wash buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol, and 25 mM imidazole) to remove unbound proteins. The protein of interest was eluted using 25 mL of elution buffer (wash buffer with 500 mM imidazole). Fractions containing the protein were pooled and further purified by gel filtration chromatography on a GE Healthcare HiLoad 16/60 Superdex 200 prep grade column pre-equilibrated with gel filtration buffer (10 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, and 5 mM DTT). Fractions containing the target protein were combined and concentrated by centrifugation in an Amicon Ultra-15 10,000 MWCO centrifugal filter unit. The final yields and concentrations were 31 mg (6 mg/mL) of Pa0148, 92 mg (13 mg/mL) of Sgx9403e, and 114 mg (13.5 mg/mL) of Sgx9403g per liter of media. Electrospray mass spectroscopy was used to obtain an accurate mass of the purified protein and establish the extent of selenomethionine labeling (Pa0148: 37430 Da/4 SeMet, Sgx9403e: 39016 Da/8 SeMet, and Sgx9403g: 39001 Da/2 SeMet).
The expression plasmids are available through the PSI Material Repository (dnasu.asu.org) as NYSGXRC clone IDs 9608a2BCt1p1, 9403e1BCt11p1, and 9403g1BCt10p1. Associated experimental information is available in the Protein Expression Purification Crystallization Database (PepcDB.pdb.org) as TargetID’s “NYSGXRC-9608a”, “NYSGXRC-9403e”, and “NYSGXRC-9403g”.
Cloning and Purification of AAur1117 from Arthrobacter aurescens TC1
AAur1117 was cloned from the genomic DNA of Arthrobacter aurescens TC1. The PCR product was amplified using the primer pair 5′-AGAGGATCCGTGGAAACTTTTGGCGAGAAAACTACC-3′ and 5′-AGAGAAGCTTTTAGGCGGGCGTAACGGAGGC-3′. The restriction sites for BamHI and HindIII were inserted into the forward and reverse primers, respectively. The PCR product was purified with a PCR clean-up system (Promega), digested with BamHI and HindIII and ligated into a pET-30a(+) vector, which was previously digested with BamHI and HindIII. The cloned gene fragment was sequenced to verify fidelity of the PCR amplification.
The gene for AAur1117 inserted in a pET-30a(+) vector was transformed into BL21(DE3) cells (Novagen). A single colony was used to inoculate a 5 mL overnight culture of LB medium containing 50 μg/mL kanamycin. Each overnight culture was used to inoculate 1.0 L of LB medium containing 50 μg/mL kanamycin. One liter cultures were grown at 37 °C and induced with 50 μM isopropyl D-thiogalactopyranoside (IPTG) when OD600 of 0.6 was reached. At the time of induction, the temperature was lowered to 20 °C and allowed to shake for 18 hours before the cells were harvested at 8,000 rpm for 10 minutes. The cells were resuspended in 50 mM HEPES buffer, pH 7.5, containing 0.1 mg/mL phenylmethylsulfonyl fluoride and 0.5 mg/mL deoxyribonuclease I from bovine pancreas. Cells were lysed by sonication. Soluble protein was separated from the cell debris by centrifugation and fractionated by ammonium sulfate precipitation. The precipitated protein (35–70% ammonium sulfate saturation) was resuspended in 50 mM HEPES buffer, pH 7.5, and loaded onto a High Load 26/60 Superdex 200 prep grade gel filtration column (GE Healthcare). Fractions containing AAur1117 were loaded onto a 6 mL ResourceQ column and eluted with a gradient of NaCl.
Crystallization and Structure Determination of Pa0148 and AAur1117
Pa0148 and AAur1117 were each concentrated to ~10 mg/ml and subjected to sitting-drop, vapor-diffusion by mixing 1 μL of protein solution and 1 μL of reservoir from Index and Crystal screen of Hampton Research. Adenine was added to the AAur1117 protein solution to 10 mM final concentration before crystallization trials were initiated. Pa0148 formed thin plate-like crystals overnight from 0.2 M magnesium chloride, 0.1 M HEPES pH 7.5, 25% w/v polyethylene glycol (PEG) 3350. AAur1117 formed rod shaped crystals over the course of three months from 0.2 M magnesium chloride hexahydrate, 0.1 M Tris hydrochloride pH 8.5, 30% w/v PEG 4000. All crystals were cryoprotected by addition of 20% glycerol (v/v) to the mother liquor and flash-frozen by direct immersion in liquid nitrogen. Pa0148 was also soaked individually in 10 mM adenine, hypoxanthine, or 6-chloropurine added to mother liquor plus 20% glycerol and flash frozen in liquid nitrogen for x-ray data collection.
For Pa0148 and AAur1117, X-ray diffraction data at the selenium absorption edge (λ=0.9795 Å) were recorded at beamlines X25 and X12C, respectively, at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. Diffraction data were processed with HKL2000 or MOSFLM/SCALA (23–25). Pa0148 crystallized in the orthorhombic space group P212121 with two molecules/asymmetric unit and diffracted to ~2.5 Å resolution. The structure of Pa0148 was determined by AUTOSOL (26), followed by autobuilding in ARP/wARP (27). Atomic models for both structures were subsequently manually adjusted using COOT (28). Structures of Pa0148 bound to adenine, hypoxanthine, or 6-chloropurine were determined via molecular replacement using PHASER (29) with the structure of Pa0148 as a search model. AAur1117 crystallized in the tetragonal space group P41212 with one molecule in the asymmetric unit, and diffracted to ~2.6 Å resolution. The structure of AAur1117 was determined via molecular replacement using the structure of Pa0148 as a search model. All five structures were refined to convergence with PHENIX (30). Data collection and refinement statistics are summarized in Table 1.
Table 1.
Data Collection and Refinement Statistics
| PDB ID | 3OU8 | 3PAO | 3PBM | 3PAN | 3RYS |
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9795 | 0.9795 | 0.9795 |
| Resolution (Å) | 50-2.51 | 50-2.40 | 50-2.60 | 50-2.63 | 50-2.60 |
| Outer shell resolution (Å) | 2.65-2.51 | 2.49-2.40 | 2.69-2.60 | 2.72-2.63 | 2.69-2.60 |
| Space group | P212121 | P212121 | P212121 | P212121 | P41212 |
| Cell dimensions | |||||
| a, b, c Cell dimensions (Å) | 44.4, 74.0, 177.9 | 44.6, 74.0, 177.0 | 44.7, 74.1, 178.2 | 44.7, 74.2, 178.0 | 124.1, 124.1, 89.7 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Number of molecules/asymmetric unit | 2 | 2 | 2 | 2 | 2 |
| Redundancy (overall/outermost shell) | 13.9(14.0) | 7.9(7.0) | 6.6 (6.3) | 14.3 (14.2) | 18.3 (18.7) |
| I/σ(I) (overall/outermost shell) | 16.2(9.8) | 18.2(5.1) | 9.28(3.32) | 19.83(8.7) | 19.83(8.7) |
| Rmerge (overall/outermost shell) | 0.126(0.223) | 0.179(0.44) | 0.176 (0.473) | 0.148 (0.333) | 0.149 (0.528) |
| Completeness (%) (overall/outermost shell) | 100.0(100.0) | 100.0(100.0) | 99.4(96.1) | 99.9(99.4) | 100.0(100.0) |
| No. of reflections | 20922 | 24150 | 19042 | 198396 | 22144 |
| Refinement statistics | |||||
| Resolution range (Å) | 50-2.51 | 50-2.40 | 50-2.60 | 50-2.63 | 50-2.60 |
| No. of reflections | 20862 | 21273 | 18038 | 17938 | 21346 |
| Completeness (work + test) (%) | 99.95 | 99.49 | 94.13 | 97.57 | 96.56 |
| Rfactor (%) | 19.04 | 18.12 | 17.26 | 19.24 | 20 |
| Rfree (%) | 25.43 | 24.03 | 23.2 | 25.28 | 25.61 |
| No. of protein atoms | 5069 | 5064 | 5064 | 5064 | 5100 |
| No. of water molecules | 177 | 176 | 133 | 148 | 85 |
| No. of ligand atoms | 2 | 22 | 22 | 22 | 22 |
| Mean B value (overall, Å2) | 18.44 | 23.13 | 20.75 | 18.57 | 21.11 |
| rmsd for bonds (Å) | 0.006 | 0.007 | 0.01 | 0.01 | 0.004 |
| rmsd for angles (°) | 0.871 | 0.991 | 1.27 | 0.701 | 0.767 |
| Ramachandran plot analysis (%) | |||||
| Most favored region (additionally allowed) | 95.02(3.53) | 95.83 (2.89) | 94.54 (4.17) | 95.83 (2.73) | 97.25 (1.83) |
| Disallowed region | 1.44 | 1.28 | 1.28 | 1.44 | 0.92 |
Activity Screens
Pa0148, AAur1117, Sgx9403e, and Sgx9403g (10 nM) were incubated for 16 hours with 2,6-diaminopurine, 6-mercaptopurine, 6-methoxypurine, 6-methylthiopurine, 6-chloropurine, 6-methylpurine, N-6-isopentyl adenine, isoguanine, N-6-methyladenine, zeatin, adenine, adenosine, 2′-deoxyadenosine, 3′-deoxyadenosine, 5′-deoxyadenosine, 2′,5′-dideoxyadenosine, AMP, ADP, ATP, S-adenosylhomocysteine, S-adenosyl methionine, N-6-methyl-2′-deoxyadenosine, 3′,5′-cyclic AMP, 5′-thiomethyladenosine, 5′-methyldeoxycytidine, cytidine, 2′-deoxycytidine, cytosine, 5-hydroxymethylcytosine, 2-chloroadenine, or 2-dimethylaminoadenine. The substrate concentration was 80 μM in each case. Enzymatic activity was monitored through changes in absorbance between 240–300 nm on a SPECTRAmax Plus spectrophotometer (Molecular Devices).
Measurement of Kinetic Constants
Assays were conducted with substrate concentrations of 3–200 μM. Deamination of adenine and 2,6-diaminopurine was monitored by following the decrease in absorbance at 262 nm and 282 nm, respectively. Dechlorination of 6-chloropurine was monitored by following the increase in absorbance at 250 nm. Formation of hypoxanthine from N-6-methyladenine and 6-methoxypurine was monitored at 270 nm. Difference extinction coefficients were calculated by subtracting the extinction coefficient of the product from the extinction coefficient of the substrate for adenine (Δε262 = 4600 M−1 cm−1), N-6-methyladenine (Δε270 = 15000 M−1 cm−1), 2,6-diaminopurine (Δε282 = 1600 M−1 cm−1), 6-chloropurine (Δε250 = 6100 M−1 cm−1) and 6-methoxypurine (Δε270 = 1900 M−1 cm−1). Production of hypoxanthine was confirmed by mass spectrometry for the reactions of adenine, 6-chloropurine and 6-methoxypurine. Deamination of 2,6-diaminopurine was confirmed using a coupled assay with guanine deaminase to produce the doubly deaminated product, xanthine. Production of methanol from 6-methoxypurine was detected using alcohol dehydrogenase (4 mg/mL) and monitoring the conversion of NAD+ to NADH at 340 nm (31).
Metal Analysis
Metal content of the proteins was determined by ICP-MS (32). Protein samples for ICP-MS were digested with HNO3 by refluxing for ~45 minutes to prevent protein precipitation during the measurement. Protein concentration was adjusted to ~1.0 μM with 1% (v/v) HNO3.
Data Analysis
Sequence alignments were created using ClustalW at http://www.compbio.dundee.ac.uk/JalviewWS/services/ClustalWS. Steady state kinetic data were analyzed using Softmax Pro, version 5.4. Kinetic parameters were determined by fitting the data to equation 1 using the nonlinear least-squares fitting program in SigmaPlot 9.0. In this equation A is the substrate concentration, Km is the Michaelis constant, v is the velocity of the reaction and kcat is the turnover number.
| (1) |
RESULTS
Protein Purification
The four enzymes that are the focus of this investigation belong to Group 3 of cog1816 (Figure 1). Pa0148 and AAur1117 were expressed in E. coli and purified to homogeneity. Pa0148 and AAur1117 contained, on average, 0.5 and 0.9 equivalents of Zn2+ per monomer. The genes encoding Sgx9403e and Sgx9403g were synthesized, expressed in E. coli, and the resulting proteins purified to homogeneity. Sgx9403e contained 0.6 equivalents of Mn2+ per monomer and Sgx9403g contained 0.8 equivalents of Mn2+ per monomer. The role of the divalent cation in the active site of adenine deaminase is to facilitate the activation of the water molecule for the hydrolytic deamination reaction (4). It is assumed that protein without a divalent cation in the active site is inactive. No attempt was made to determine the dissociation constant for the protein-metal complex.
Three-Dimensional Structures of Pa0148 and AAur1117
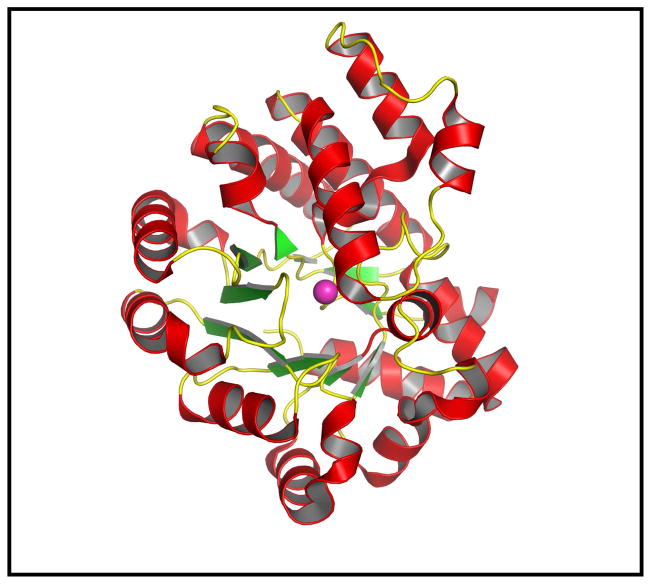
Pa0148 and AAur1117 were crystallized and their three-dimensional structures determined to resolution limits of ~2.5 and ~2.6 Å, respectively. A ribbon diagram illustrating the overall protein fold for Pa0148 is presented in Figure 2. The sequence identity of these two proteins is 53% and the RMSD for the equivalent α-carbon atoms is 0.67 Å. The two proteins fold into a distorted (β/α)8-barrel with a single divalent cation bound in the active site. The ligand-free structure of Pa0148 (PDB code: 3OU8) reveals those residues responsible for the coordination of zinc in the active site and the residues that stabilize the position of a metal-bound water molecule. Metal binding residues for Pa0148 include an HxH motif from the C-terminus of β-strand 1 (His-16 and His-18), a histidine from the C-terminus of β-strand 5 (His-196) and an aspartate from the C-terminus of β-strand 8 (Asp-277). The presumed hydrolytic water molecule is located 3.6 Å from the zinc ion. This water molecule is 3.6 Å from the side chain of His-220 and 3.0 Å from the side chain of Asp-277. The metal coordination scheme for Pa0148 is presented in Figure 3.
Figure 2.
Three dimensional structure of Pa0148 (PDB code: 3OU8). The eight β-strands that form the core (β/α)8-barrel are shown in green and the α-helices are red. The single zinc cation is shown in pink.
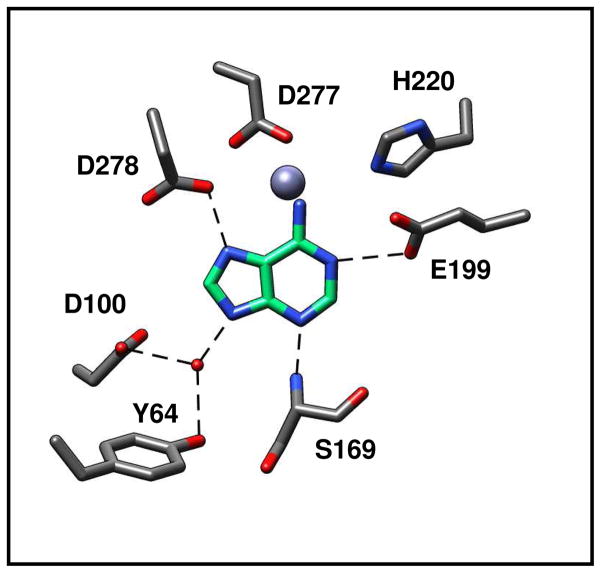
Figure 3.
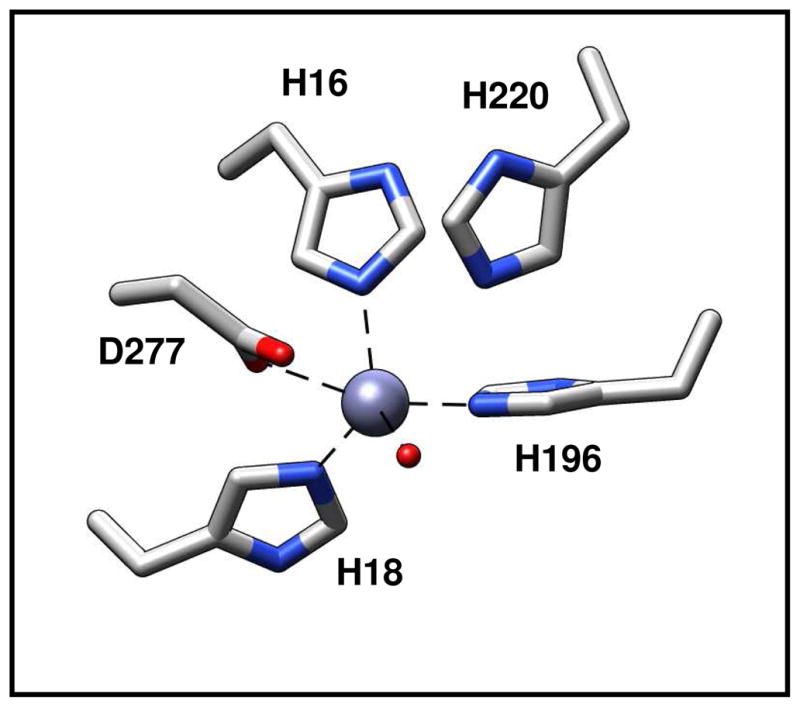
Active site of Pa0148 showing the residues that coordinate the divalent metal ion. The divalent cation is presented as a grey sphere and the water molecule is shown in red.
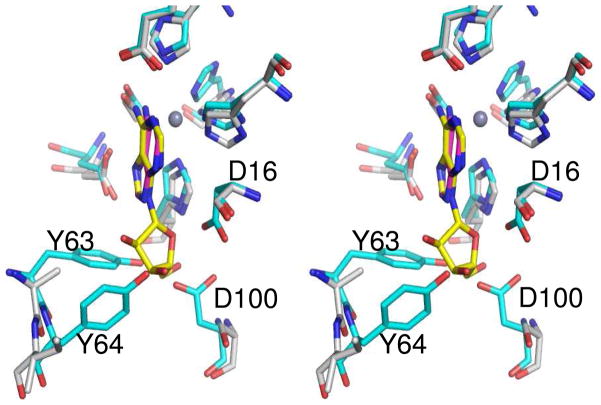
Co-crystal structures of Pa0148 bound to adenine (PDB code: 3PAO), hypoxanthine (PDB code: 3PAN), and 6-chloropurine (PDB code: 3PBM) were obtained by soaking pre-formed Pa0148 apo-protein crystals. Adenine binding to Pa0148 is mediated by Tyr-64 and Asp-100 (Figure 4). These two residues hydrogen bond to a water molecule that is, in turn, hydrogen bonded to N9 of the purine ring. N3 of the purine ring hydrogen bonds to the backbone amide group of Ser-169. Additional intermolecular interactions include Asp-278 to N7 and Glu-169 to N1 of the purine ring. The putative hydrolytic water molecule is not observed in the active site of Pa0148 when either the substrate or products are bound in the active site. The structure of Pa0148 with hypoxanthine bound in the active site is nearly identical to the structure with adenine (RMSD = 0.11 Å) except that the substrate is rotated slightly. A similar rotation is apparent in the structure of 6-chloropurine bound to Pa0148; the RMSD between the two structures is 0.1 Å. An overlay of the active sites showing the relative positions of adenine and hypoxanthine bound to Pa0148 is presented in Figure 5. No conformational changes to the protein are apparent upon the binding of either substrate or products. The co-crystal structure of AAur1117 complexed with adenine (PDB: 3RYS) demonstrates a binding mode similar to those of 6-chloropurine and hypoxanthine in the active site of Pa0148.
Figure 4.
Active site of Pa0148 showing the residues and water molecule (red) that interaction with the substrate adenine.
Figure 5.
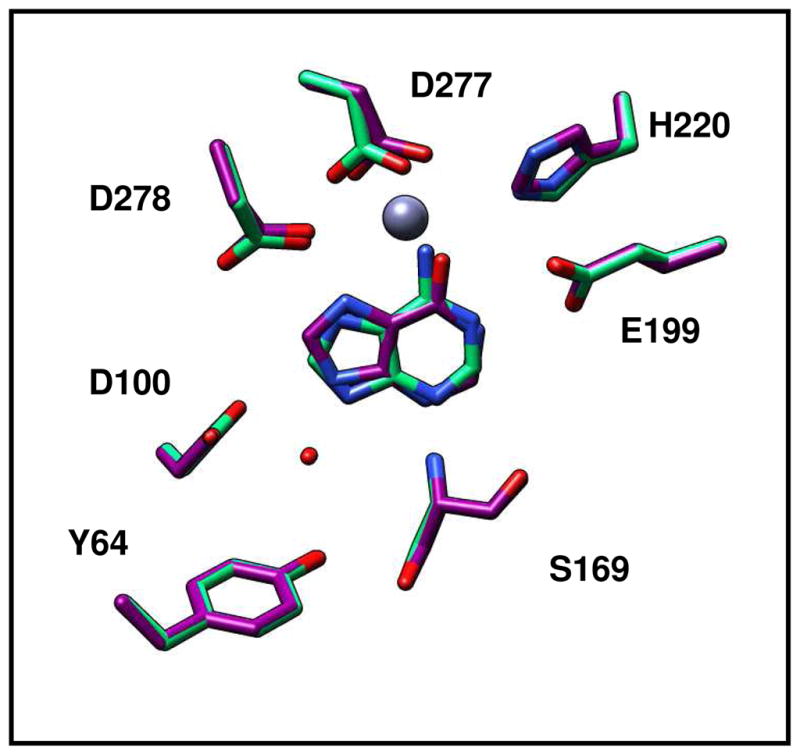
Active site of Pa0148 showing a comparison of the binding of the substrate adenine (PDB code: 3PAO) and the product hypoxanthine (PDB code: 3PAN). The divalent cation is presented as a grey sphere and the water molecule is shown in red.
Substrate Specificity
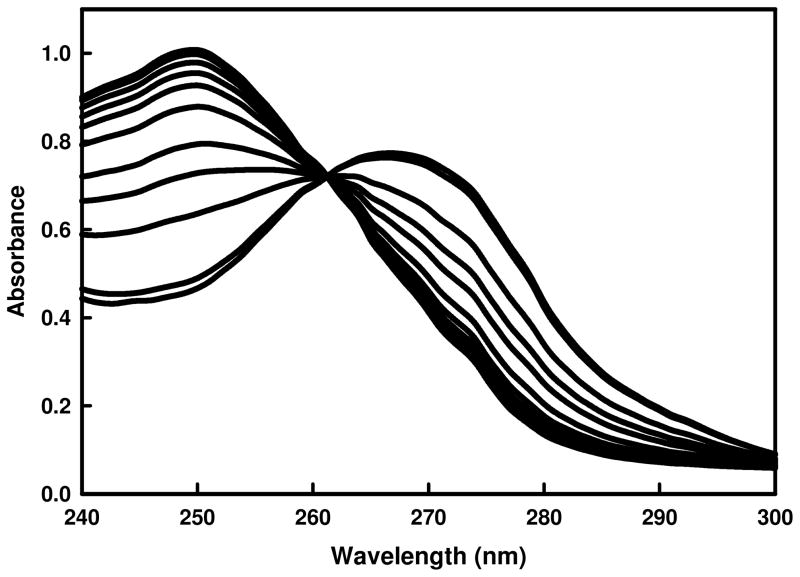
The substrate profiles for Pa0148, AAur1117, Sgx9403e, and Sgx9403g were determined by monitoring changes in absorbance between 240–300 nm after the addition of enzyme to a library of modified purines and other nucleic acid derivatives. The best substrates for these four enzymes are adenine, 6-chloropurine, N-6-methyladenine, 6-methoxypurine, and 2,6-diaminopurine. All four enzymes were unable to deaminate adenosine. The upper limit for the deamination of adenosine, relative to adenine by Pa0148 is 0.01%. The change in absorbance as 6-chloropurine is converted to hypoxanthine in the presence of Pa0148 is shown in Figure 6. The absorbance decreases at 267 nm and increases at 250 nm. Production of hypoxanthine was confirmed by mass spectrometry for the reactions of adenine, 6-chloropurine and 6-methoxypurine. A coupled assay with alcohol dehydrogenase was used to detect the formation of methanol from 6-methoxypurine by monitoring the conversion of NAD+ to NADH at 340 nm. The deamination of 2,6-diaminopurine was confirmed using a coupled assay with guanine deaminase for the ultimate formation of xanthine via guanine. The kinetic constants for the reactions of adenine, 6-chloroadenine, N-6-methyladenine, 6-methoxypurine, and 2,6-diaminopurine are presented in Table 2. Adenine was the best overall substrate for all four enzymes examined, with values of kcat/Km exceeding 105 M−1s−1.
Figure 6.
Time course for the changes in absorbance as Pa0148 dechlorinates 6-chloropurine to hypoxanthine. The decrease the absorbance at 267 nm is coupled with an increase in the absorbance at 250 nm as 6-chloropurine is converted to hypoxanthine.
Table 2.
Kinetic Constants for Pa0148, AAur_1117, Sgx9403e and Sgx9403ga
| Enzyme | Adenine | 6-chloropurine | N-6-methyladenine | 2,6-diaminopurine | 6-methoxypurine | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (M−1s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1s−1) | |
| Pa0148 | 36 | 25 | 1.4×106 | 1.9 | 260 | 7.3×103 | 1.28 | 88 | 1.5×104 | 17 | 140 | 1.2 ×105 | 7.1 | 270 | 2.6×104 |
| AAur_1117 | 61 | 200 | 3.1×105 | - | - | 9.6×103 | - | - | 1.3×103 | 6.9 | 290 | 2.4×104 | 1.9 | 160 | 1.2×104 |
| Sgx9403e | 62 | 200 | 3.1×106 | 0.84 | 56 | 1.5×104 | 0.48 | 90 | 5.3×103 | 1.8 | 70 | 2.8×104 | 0.52 | 130 | 4.0×103 |
| Sgx9403g | 56 | 170 | 3.3×105 | 0.42 | 26 | 1.6×104 | - | - | 1.6×103 | - | - | 1.9×104 | 2.2 | 140 | 1.7×104 |
Standard errors on the kinetic constants are less than 20%.
DISCUSSION
Substrate Specificity
We have identified a large group of bacterial enzymes from cog1816 within the amidohydrolase superfamily of enzymes that are currently incorrectly annotated as adenosine deaminases. The best substrate for this group of enzymes identified thus far is adenine. Additional substrates include 2,6-diaminopurine, 6-chloropurine, 6-methoxypurine, and N-6-methyladenine (Scheme 1). These enzymes show no activity with adenosine. The ability to accept an amino substituent attached to C2 (for example, 2,6-diaminopurine) is similar to that of the authentic adenosine deaminases from cog1816, which will accept a methyl or amino group at this position, but not a halide substituent (33). The 2-amino group is apparently able to fit into a pocket within the active site that contains two water molecules in the crystal structure of Pa0148 and AAur1117. Hydrogen bonding interactions between the waters and the amino group help to stabilize the binding of 2,6-diaminopurine, but apparently do not allow for substitution of a halide in this position. 2,6-Diaminopurine has been used in the treatment of cancer (34) and at one time was proposed to be an intermediate in the conversion of guanine to adenine in mammals (35). More recently, 2,6-diaminopurine has been shown to bind to purine riboswitches (36, 37).
Scheme 1.
The binding pocket near the N6 amino group of adenine in the crystal structure of Pa0148 is large enough to accommodate an additional methyl group, allowing for the binding and turnover of N-6-methyladenine and 6-methoxypurine. This observation is in agreement with the reported substrate specificity of the mammalian adenosine deaminase (38) and that of E. coli adenosine deaminase from cog1816 (data not shown), both of which accept the N-6-methyl and 6-methoxy derivatives of adenosine as substrates. N-6-methyladenine is a naturally occurring base found in 1–2% of the total adenine content of bacterial genomes (39). Methylation of adenine serves to protect host DNA from endonucleases which target foreign DNA and regulates the interaction between DNA and some DNA-binding proteins (39, 40).
Deamination of N-6-methyladenine has been observed in cell free extracts from E. coli (41) and an enzyme has been identified with this catalytic activity from B. halodurans (5). However, the N-6-methyladenine deaminase (6-MAD) from B. halodurans (Bh0637) is found in cog1001 from the amidohydrolase superfamily. The 6-MAD enzymes from cog1001 have a significantly higher activity with N-6-methyl adenine than with adenine. With the Group 3 enzymes from cog1816, adenine is deaminated about 2-orders of magnitude faster than N-6-methyladenine.
Sequence Analysis
The four enzymes analyzed in this investigation share sequence identity of more than 50% to one another (Figure 7). Compared to adenosine deaminase from E. coli (locus tag: b1623) from group 5 of cog1816, the sequence identity is less than 30%. All of the residues that coordinate the divalent cation in the active site are conserved among the Group 5 and Group 3 enzymes, including the HxH motif following β-strand 1, a histidine at the C-terminus of β-strand 5 and an aspartate at the C-terminus of β-strand 8. These residues are highlighted in red in Figure 7. Additional conserved residues include an aspartate that follows the invariant aspartate which is coordinated to the divalent cation found at the C-terminal end of β-strand 8. This residue hydrogen bonds to N7 of the adenine ring. Also conserved among the Group 3 and Group 5 enzymes of cog1816 is the glutamate from the HxxE motif at the C-terminus of β-strand 5. This residue is responsible for the protonation of N1 during the deamination reaction. The other functionally important residue is the histidine at the C-terminus of β-strand 6 that appears to form a hydrogen bond with the putative hydrolytic water and may participate in proton transfer reactions. These residues are highlighted in blue in Figure 7.
Figure 7.
Multiple-sequence alignment of an adenosine deaminase from E. coli Str. K12 (b1623), and four adenine deaminases: Pa0148, AAur_1117, Sgx9403e, and Sgx9303g. The eight β-strands are highlighted with a light grey background. Residues highlighted in red coordinate the active site metal ion. Those residues highlighted in blue are conserved in Groups 3 and 5 of cog1816 and are involved in catalysis or recognition of the adenine portion of the substrate. Residues highlighted in green are only conserved in Group 3 of cog1816 and these residues function in defining the substrate specificity.
Pa0148 and the other members of Group 3 of cog1816 share a four residue deletion between β-strands 1 and 2. In addition, Group 3 proteins possess a conserved tyrosine repeat between β-strands 1 and 2. This pair of tyrosine residues in the crystal structures of Pa0148 and AAur1117 occurs within the last of three α-helices connecting β-strands 1 and 2. The second tyrosine, together with an aspartate from the C-terminal end of β-strand 4, is responsible for positioning a water molecule to hydrogen bond with N3 of adenine. Adenosine is apparently unable to bind in the active site of Group 3 proteins because the first tyrosine occludes ribose binding (see Figure 8) (17). A further impediment to ribose binding is an aspartate residue at the C-terminus of β-strand 2 which occupies the binding site of the 5′-hydroxyl group of adenosine for the Group 5 adenosine deaminases. In Pa0148 this residue is Asp-168. Adenine binding residues are highlighted in green in Figure 7. The four residue deletion, the two conserved tyrosines following β-strand 1, the aspartate following β-strand 2 and the aspartate from β-strand 4 are fully conserved in Pa0148 and approximately 120 other proteins from Group 3 of cog1816. We conclude that these enzymes are adenine deaminases, not adenosine deaminases, as currently annotated. These proteins are listed in Table S1 in the Supplementary Information. These adenine deaminases are similar in amino acid sequence to a previously described fungal adenine deaminases (42).
Figure 8.
Stereoview of an overlay between Pa0148 with adenine bound (PDB code: 3PAO) and adenosine deaminase from human malaria parasite with bound adenosine (PDB code: 2PGF) at the active site. Pa0148 and adenosine deaminase active site residues are colored in cyan and grey, respectively. Adenine and adenosine are colored in magenta and yellow, respectively. The Zn2+ from Pa0148 colored in dark grey.
Comparison to Adenine Deaminase from cog1001
The prototypical adenine deaminase from E. coli (locus tag: b3665) is also a member of the amidohydrolase superfamily but belongs to cog1001. The E. coli adenine deaminase has a binuclear metal center with a high sensitivity to iron and will apparently accept no substrate other than adenine (43). The active site of the E. coli adenine deaminase resembles the active sites of urease and phosphotriesterase. It is unclear why some bacteria possess a mononuclear adenine deaminase and others from the same class of bacteria possess a binuclear adenine deaminase. The adenine deaminases from cog1001 and cog1816 have nearly equal representation among the bacterial genomes. Although there is overlap between the types of bacteria that possess a specific adenine deaminase, each species of bacteria appears to possess only one adenine deaminase.
Strategy for Functional Annotation
The COG database was used to construct a sequence similarity network of cog1816, which identified enzymes belonging to Group 3 as significantly different from experimentally verified adenosine deaminases in Group 5. This divergence and the absence of key residues in sequence alignments between proteins in Groups 3 and 5, lead to the prediction that proteins within Group 3 would have a unique substrate profile, other than the deamination of adenosine. After the structure for Pa0148 was determined, modeling of adenosine in the active site of this protein demonstrated that the ribose binding region was obscured by residues found only in proteins of Group 3. This observation prompted the successful search for the true substrate for Pa0148 and related proteins, using adenine and modified purines. The subsequent determination of the structure of Pa0148 in the presence of adenine and hypoxanthine illustrated how these compounds are recognized within the active site. A similar inspection of the other major groups within cog1816 indicate that they almost certainly catalyze the deamination of substrates other than adenosine.
Supplementary Material
Acknowledgments
We gratefully acknowledge data collection support from National Synchrotron Light Source. We also thank the NYSGXRC protein production team for preparation of Pa0148, Sgx9403e and Sgx9403g.
ABBREVIATIONS
- ADE
adenine deaminase
- ADD
adenosine deaminase
- COG
cluster of orthologous groups
- IPTG
isopropyl-β-galactoside
- DTT
dithiothreitol
- AHS
amidohydrolase superfamily
- ICP-MS
inductively coupled plasma mass spectrometry
- 6-MAD
N-6-methyladenine deaminase
Footnotes
This work was supported in part by National Institutes of Health (GM 71790 and GM 74945) and the Robert A. Welch Foundation (A-840). The X-ray coordinates and structure factors have been deposited in the Protein Data Bank (PDB: 3OU8, 3PAN, 3PAO, 3PBM and 3RYS)
A list of proteins from cog1816 that are predicted to catalyze the deamination of adenine is provided (Table S1). This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 2.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;28:72–82. [PubMed] [Google Scholar]
- 4.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 5.Kamat SS, Fan H, Sauder JM, Burley SK, Shoichet BK, Sali A, Raushel FM. Enzymatic deamination of the epigenetic base N-6-methyladenine. J Am Chem Soc. 2011;133:2080–2083. doi: 10.1021/ja110157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall RS, Agarwal R, Hitchcock D, Sauder JM, Burley SK, Swaminathan S, Raushel FM. Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase. Biochemistry. 2010;49:4374–4382. doi: 10.1021/bi100252s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Quiocho FA. Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity. Biochemistry. 1998;37:8314–8324. doi: 10.1021/bi980324o. [DOI] [PubMed] [Google Scholar]
- 8.Ho MC, Cassera MB, Madrid DC, Ting LM, Tyler PC, Kim K, Almo SC, Schramm VL. Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases. Biochemistry. 2009;48:9618–9626. doi: 10.1021/bi9012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baer HP, Drummond GI, Gillis J. Studies on the specificity and mechanism of action of adenosine deaminase. Arch Biochem Biophys. 1968;123:172–178. doi: 10.1016/0003-9861(68)90116-1. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita T, Nakanishi I, Terasaka T, Kuno M, Seki N, Warizaya M, Matsumura H, Inoue T, Takano K, Adachi H, Mori Y, Fujii T. Structural basis of compound recognition by adenosine deaminase. Biochemistry. 2005;44:10562–10569. doi: 10.1021/bi050529e. [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita T, Tada T, Nakanishi I. Conformational change of adenosine deaminase during ligand-exchange in a crystal. Biochem Biophys Res Commun. 2008;373:53–57. doi: 10.1016/j.bbrc.2008.05.180. [DOI] [PubMed] [Google Scholar]
- 12.Koch AL, Vallee G. The properties of adenosine deaminase and adenosine nucleoside phosphorylase in extracts of Escherichia coli. J Biol Chem. 1959;234:1213–1218. [PubMed] [Google Scholar]
- 13.Lin J, Westler WM, Cleland WW, Markley JL, Frey PA. Fractionation factors and activation energies for exchange of the low barrier hydrogen bonding proton in peptidyl trifluoromethyl ketone complexes of chymotrypsin. Proc Natl Acad Sci U S A. 1998;95:14664–14668. doi: 10.1073/pnas.95.25.14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gleeson MP, Burton NA, Hillier IH. Prediction of the potency of inhibitors of adenosine deaminase by QM/MM calculations. Chem Commun (Camb) 2003:2180–2181. doi: 10.1039/b305790a. [DOI] [PubMed] [Google Scholar]
- 15.Sadat Hayatshahi SH, Abdolmaleki P, Ghiasi M, Safarian S. QSARs and activity predicting models for competitive inhibitors of adenosine deaminase. FEBS Lett. 2007;581:506–514. doi: 10.1016/j.febslet.2006.12.050. [DOI] [PubMed] [Google Scholar]
- 16.Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 17.Larson ET, Deng W, Krumm BE, Napuli A, Mueller N, Van Voorhis WC, Buckner FS, Fan E, Lauricella A, DeTitta G, Luft J, Zucker F, Hol WG, Verlinde CL, Merritt EA. Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity. J Mol Biol. 2008;381:975–988. doi: 10.1016/j.jmb.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss PM, Cook PF, Hermes JD, Cleland WW. Evidence from nitrogen-15 and solvent deuterium isotope effects on the chemical mechanism of adenosine deaminase. Biochemistry. 1987;26:7378–7384. doi: 10.1021/bi00397a027. [DOI] [PubMed] [Google Scholar]
- 19.Sideraki V, Mohamedali KA, Wilson DK, Chang Z, Kellems RE, Quiocho FA, Rudolph FB. Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase. Biochemistry. 1996;35:7862–7872. doi: 10.1021/bi952920d. [DOI] [PubMed] [Google Scholar]
- 20.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamedali KA, Kurz LC, Rudolph FB. Site-directed mutagenesis of active site glutamate-217 in mouse adenosine deaminase. Biochemistry. 1996;35:1672–1680. doi: 10.1021/bi9514119. [DOI] [PubMed] [Google Scholar]
- 22.Sideraki V, Wilson DK, Kurz LC, Quiocho FA, Rudolph FB. Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation. Biochemistry. 1996;35:15019–15028. doi: 10.1021/bi961427e. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography. 1992:26. [Google Scholar]
- 25.4CCPN. The CCP4 suite: programs for protein crystallography. Acta Crystallography D. 1994:50. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Hattne J, Lamzin VS. Pattern-recognition-based detection of planar objects in three-dimensional electron-density maps. Acta Crystallogr D Biol Crystallogr. 2008;D64:834–842. doi: 10.1107/S0907444908014327. [DOI] [PubMed] [Google Scholar]
- 27.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 31.Bergmeyer HU, editor. Methods of Enzymatic Analysis. Academic Press; New York, NY: 1974. [Google Scholar]
- 32.Hall RS, Xiang DF, Xu C, Raushel FM. N-Acetyl-D-glucosamine-6-phosphate deacetylase: substrate activation via a single divalent metal ion. Biochemistry. 2007;46:7942–7952. doi: 10.1021/bi700543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokozeki K, Tsuji T. A novel enzymatic method for the production of purine-2′-deoxyribonucleosides. J Mol Catal B-Enzym. 2000;10:207–213. [Google Scholar]
- 34.Burchenal JH, Karnofsky DA, Kingsley-Pillers EM, Southam CM, Myers WP, Escher GC, Craver LF, Dargeon HW, Rhoads CP. The effects of the folic acid antagonists and 2,6-diaminopurine on neoplastic disease, with special reference to acute leukemia. Cancer. 1951;4:549–569. doi: 10.1002/1097-0142(195105)4:3<549::aid-cncr2820040308>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 35.Bendich A, Furst SS, Brown GB. On the role of 2,6-diaminopurine in the biosynthesis of nucleic acid guanine. J Biol Chem. 1950;185:423–433. [PubMed] [Google Scholar]
- 36.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J Am Chem Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 38.Orsi BA, McFerran N, Hill A, Bingham A. Kinetics and the mechanism of action of adenosine aminohydrolase. Biochemistry. 1972;11:3386–3392. doi: 10.1021/bi00768a011. [DOI] [PubMed] [Google Scholar]
- 39.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remy CN. Metabolism of 6-methylaminopurine: synthesis and demethylation by Escherichia coli. J Biol Chem. 1961;236:2999–3005. [PubMed] [Google Scholar]
- 42.Ribard C, Rochet M, Labedan B, Daignan-Fornier B, Alzari P, Scazzocchio C, Oestreicher N. Sub-families of alpha/beta barrel enzymes: a new adenine deaminase family. J Mol Biol. 2003;334:1117–1131. doi: 10.1016/j.jmb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Kamat SS, Bagaria A, Kumaran D, Holmes-Hampton GP, Fan H, Sali A, Sauder JM, Burley SK, Lindahl PA, Swaminathan S, Raushel FM. Catalytic Mechanism and Three-Dimensional Structure of Adenine Deaminase. Biochemistry. 2011;50:1917–1927. doi: 10.1021/bi101788n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.