Abstract
The extent to which the transition to agriculture in Europe was the result of biological (demic) diffusion from the Near East or the adoption of farming practices by indigenous hunter–gatherers is subject to continuing debate. Thus far, archaeological study and the analysis of modern and ancient European DNA have yielded inconclusive results regarding these hypotheses. Here we test these ideas using an extensive craniometric dataset representing 30 hunter–gatherer and farming populations. Pairwise population craniometric distance was compared with temporally controlled geographical models representing evolutionary hypotheses of biological and cultural transmission. The results show that, following the physical dispersal of Near Eastern/Anatolian farmers into central Europe, two biological lineages were established with limited gene flow between them. Farming communities spread across Europe, while hunter–gatherer communities located in outlying geographical regions adopted some cultural elements from the farmers. Therefore, the transition to farming in Europe did not involve the complete replacement of indigenous hunter–gatherer populations despite significant gene flow from the Southwest Asia. This study suggests that a mosaic process of dispersal of farmers and their ideas was operating in outlying regions of Europe, thereby reconciling previously conflicting results obtained from genetic and archaeological studies.
Keywords: Neolithic transition, demic diffusion, cultural diffusion, craniometrics, demography
1. Introduction
The transition to agriculture in Europe has been subject to intense debate for over a century, with the major controversy centring on the extent to which it involved the acculturation of indigenous (Mesolithic) hunter–gatherer populations [1] or the replacement of hunter–gatherers by (Neolithic) farmers dispersing from the Near East [2]. These contrasting demographic models are often referred to as the cultural diffusion and demic diffusion models, respectively [2,3]. Archaeological studies of Mesolithic and Neolithic sites [4] and analyses of modern European genetic variation [5–12] have yielded diverse and often conflicting conclusions regarding this process. Recent ancient DNA studies have also provided inconsistent results regarding the biological relationship between Mesolithic and Neolithic populations [13–19], suggesting that the demographic transition was not uniform across Europe but rather represents a mosaic of population replacement, admixture and adoption of farming practices by indigenous populations. Not all genetic haplotypes found in ancient Mesolithic and Neolithic populations survive in modern European populations, indicating that post-Neolithic migrations and expansions have diluted the genetic legacy of earlier populations (cf. [15]).
It has been demonstrated [3] that in the case of central and southeast Europe, Mesolithic and Neolithic craniometric data support a model of demic diffusion. These results are in agreement with the archaeological record which attests that farming emerges in central Europe in the form of a ‘Neolithic package’, including pottery and a subsistence strategy reliant on domesticates originating in the Near East/Anatolia (e.g. [20]). Thus, the best fit model for the initial transition to agriculture in Europe is one of demic diffusion from Southwest Asia, rather than the gradual adoption of Neolithic economy and culture by local hunter–gatherer groups (e.g. [3,7,11,19,21]).
Conversely, the transition to farming in the outlying regions of Europe was probably a more complex and lengthy process involving acculturation of local hunter–gatherer groups [22–26]. The archaeological record of these areas indicates a deceleration in the spread of farming, which may have been owing to higher population densities of Late Mesolithic hunter–gatherers [22,24]. Additionally, seasonal differences in climate, poor soils and dense forest were perhaps not favourable for a subsistence strategy originating in the semi-arid regions of Southwest Asia. A frontier between fully agricultural societies such as the Linienbandkeramik (LBK) and late hunter–gatherer groups could have resulted in a long ‘substitution’ phase whereby hunter–gatherers selectively integrated some cultigens and domestic livestock into their subsistence base, as well as began to use pottery [22,26]. In southern Scandinavia, the archaeological evidence strongly supports a model of indigenous transition of late hunter–gatherer Ertebølle to a fully Neolithic TRB (Funnel Beaker Culture) economy [24], although analysis of ancient DNA tells a different story [17]. In the circum-Baltic area, a number of ‘Forest Neolithic’ cultures emerged during the seventh millennium BP, continuing the Mesolithic hunting–gathering–fishing lifestyle by incorporating wild fauna and edible plant species into their diets but also living in semi-permanent locales [23,26].
Here, using a large metric dataset of 542 Mesolithic and Neolithic crania, we test whether the spread of agriculture to outlying regions (particularly the circum-Baltic region) followed a model of population dispersal or the adoption of farming practices by indigenous Mesolithic populations. There is now a growing body of literature (e.g. [27–32]) demonstrating that global human cranial variation is best explained by a neutral microevolutionary model of mutation and genetic drift [33]. These studies employ numerous craniometric datasets and different analytical approaches, yet all show that modern human cranial form can be considered an accurate proxy for neutral genetic information regarding population history. Studies of modern European DNA are affected by post-Neolithic gene flow and local population extinctions and, hence, simply cannot capture the relevant genetic variation. While ancient DNA studies directly sample the populations of interest, the resultant datasets are, at present, not geographically or chronologically detailed enough to model continent-wide processes. As such, the use of craniometric data offers a unique and valuable alternative means of quantifying and modelling this complex demographic shift.
2. Material and methods
(a). Materials
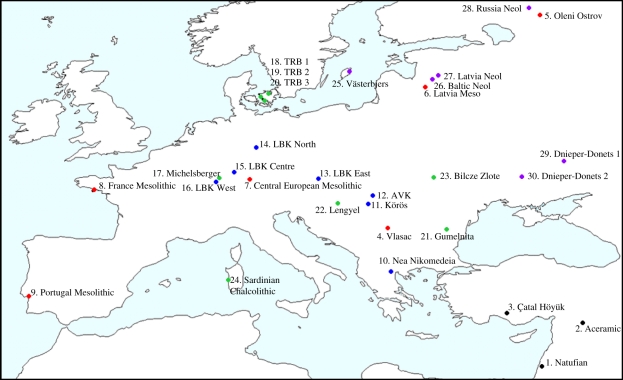
A dataset of 15 standard craniometric variables taken on 542 crania was compiled. These samples were divided into 30 operational taxonomic units (OTUs) (electronic supplementary material, table S1), each comprising at least 10 individuals. In all cases, an OTU comprised specimens from a single major archaeological phase. Whenever possible, OTUs were constructed using specimens from a single site (e.g. Çatal Höyük, Bilcze Zlote) or specimens from a well defined phase in a given region (e.g. LBK East). Sampling was constrained by uneven spatial, temporal and archaeological representativeness of certain phases, yet this dataset comprises the best available cranial samples whose archaeological contexts and skeletal preservation facilitate their inclusion in these OTUs. Fifteen of the 30 OTUs were previously employed in the study by Pinhasi & von Cramon-Taubadel [3], thus the present dataset includes more extensive geographical (Scandinavia, the Baltic region, Eastern Europe) and temporal (11 500–1500 BC) coverage (figure 1).
Figure 1.
Geographical distribution of 30 OTUs employed.
(b). Craniometric distance matrix
The craniometric data comprised 15 standard calliper measurements (electronic supplementary material, table S2) taken on each skull. Given the fragmentary nature of many of these archaeological specimens, some of the data were missing from the initial database. Only crania with data present for at least 10 out of the 15 (i.e. 70%) measurements were included in the analysis [34]. Missing data were estimated in SPSS v. 16 using multiple linear regressions, within-sexes and using the specimens with a complete set of measurements within each OTU. Sex was determined on the basis of morphological traits of the pelvis (in all cases in which at least one of the os coxae was preserved) or on the basis of the skull using standard anthropological methods [35–37]. These data were then adjusted for isometric scaling by dividing each cranial variable by the geometric mean of all variables for that individual [38,39]. A craniometric distance matrix (D-matrix) was computed for all 30 OTUs under the assumption of complete heritability (i.e. h2 = 1) using the Relethford & Blangero [40] estimator of population genetic affinities based on quantitative traits. The conservative choice of h2 = 1 (i.e. minimum genetic distance) was made given the lack of population-specific heritability estimates, but the choice of heritability value does not impact the proportionate structure of the resultant D-matrix or, therefore, the results of the Mantel tests.
Two methods were employed to visualize the major craniometric affinity patterns among the OTUs. First, a principal coordinates analysis of the resulting D-matrix was conducted and thereafter, the craniometric distance matrix was subject to two linkage methods, minimum evolution [41] and neighbour-joining [42], to generate phenetic tree topologies. While unrooted trees were generated in each case, the trees were visualized as rooted by the oldest OTU (Natufian) for visual comparability. The software Mega v. 3.1 [43] was used to generate trees, and the software Treeview v. 1.6.6 [44] was employed to modify tree figures.
(c). Hypothetical models
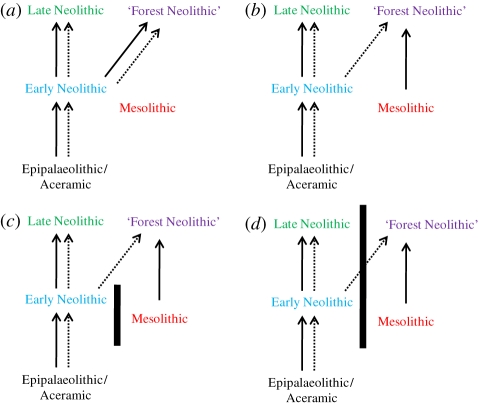
In order to construct alternative models for the Neolithization process in Europe, OTUs were organised into five broad categories represented by different colours in figure 1: (i) three Early Neolithic/Epipalaeolithic OTUs from the Near East and Anatolia (black), (ii) six Mesolithic OTUs from across Europe (red), (iii) seven Early (older than 5000 BC) Neolithic OTUs (blue), (iv) eight Late (younger than 5000 BC) Neolithic OTUs (green), and (v) six Late Forest Neolithic OTUs (purple).
In contrast with previous studies (e.g. [45]) we did not employ arbitrary values to model hypothesized biological distances between OTUs. Instead, we created a series of alternative models based on the evolutionary expectation that neutral (cranial) variation will follow a pattern of isolation-by-geographical and temporal distance (IBD) [28,46]. A strong linear relationship between geographical distance and neutral genetic/craniometric distance has been shown previously (e.g. [28]). Thus, in the absence of natural selection or disruptive long-range dispersal, craniometric distance is expected to be strongly correlated with geographical distance. However, the effects of temporal variation must also be controlled for [46]. On the basis of work by Konigsberg [46,47], there is empirical evidence of temporal autocorrelation for craniometric data, although the exact nature of this relationship is not known. Moreover, it has been shown that under a neutral microevolutionary model, we would expect a positive significant relationship between craniometric and geographical distance, once temporal distance is controlled for [46]. Therefore, in each analysis, we compared the single craniometric D-matrix with alternative models based on pairwise geographical distances between OTUs, while simultaneously controlling for the temporal distance between OTUs [3].
A null (neutral) model was constructed by calculating the pairwise great circle distances in kilometres between all OTUs [48]. The null model represents pure cultural diffusion, whereby the spread of agriculture to Europe is the result of the acculturation of indigenous populations (i.e. the spread of farming ideas, rather than farmers). However, the null model is unrealistic, as it has previously been shown [3] that the Early Neolithic OTUs from central Europe share close cranial affinities with Early Neolithic OTUs from Southwest Asia, congruent with a model of demic diffusion into central Europe. Therefore, the null geographical model was subsequently altered to reflect the four alternative scenarios (Models 1–4) depicted in figure 2. Because of the need to choose arbitrary distance values to increase/decrease the craniometric affinities between pairs of OTUs, all hypothesized dispersal events were modelled as changing by 500, 1000 and 1500 km in order to investigate the effect of using different quantities. A minimum distance of 500 km was chosen as it has been noted [20] that distances of the order of at least 300–500 km should be employed to detect true instances of dispersal rather than inherent ‘noise’ owing to the resolution of the archaeological record. All hypothesized barriers to gene flow between OTUs were conservatively modelled as being relatively weak (500 km increase).
Figure 2.
Four alternative hypothetical models describing the biological and cultural relationships between OTUs. Solid arrows, biological transmission; dashed arrows, cultural transmission; solid lines, barrier to gene flow. (a) Model 1; (b) Model 2; (c) Model 3; (d) Model 4.
Model 1 (figure 2a): pairwise distances between Near Eastern OTUs ‘Aceramic’ and ‘Çatal Höyük’ and all later OTUs were reduced to reflect the effects of the initial demic diffusion event from the Near East. This scenario hypothesizes that Mesolithic populations were completely replaced by incoming farmers and, therefore, left no biological descendents. In addition, Forest Neolithic are considered biologically equivalent to all other ‘true’ Neolithic groups. Thus, Model 1 suggests that, while the initial transition to agriculture was the result of demic diffusion, subsequent diffusion of the Neolithic across Europe was purely cultural.
Model 2 (figure 2b): pairwise distances between Near Eastern OTUs Aceramic and Çatal Höyük and all true Neolithic OTUs were reduced to reflect the effects of the initial demic diffusion from the Near East. Pairwise distances between Mesolithic and Forest Neolithic are reduced suggesting a stronger ancestor–descendant relationship. Thus, Model 2 suggests that Forest Neolithic groups (purple) are actually acculturated hunter–gatherer populations descended from earlier Mesolithic groups (red), while late ‘true’ Neolithic OTUs (green) are the biological descendents of early Neolithic (blue) populations.
Model 3 (figure 2c) is the same as Model 2 but with an increase in the pairwise distances between contemporaneous Mesolithic and Early Neolithic reflecting a barrier to normal gene flow following the demic diffusion from the Near East. While both Models 1 and 2 differ in terms of the biological relationships between early and late groups, ‘background’ gene flow is occurring between all OTUs consistent with a neutral model of isolation-by-distance. By contrast, Model 3 considers that the transition to agriculture created a cultural barrier between contemporaneous Mesolithic (red) and early Neolithic (blue) populations. Subsequently, following the adoption of Neolithic cultural elements by indigenous Forest Neolithic (purple), this gene flow barrier becomes relaxed such that contemporaneous ‘true’ and Forest Neolithic populations are freely admixing.
Model 4 (figure 2d) is the same as Model 3, but with the barrier to gene flow extending through time between contemporaneous Late true and Forest Neolithic populations. Model 4 is, therefore, a more extreme version of Model 3, whereby the cultural barrier to gene flow is continuous through time, creating two distinct lineages; the red/purple indigenous lineage and the blue/green Neolithic lineage. Despite the adoption of some cultural practices by the Forest Neolithic, they remain a biologically distinct group separate from their contemporaneous ‘true’ Neolithic neighbours.
The temporal distance matrix was calculated as the difference in the average age (in years) of all pairs of OTUs (electronic supplementary material, table S1).
(d). Analytical methods
The congruence of the craniometric D-matrix and each of the five (null plus four alternative) geographical matrices was quantified using partial Mantel matrix tests [49], and controlling for temporal distance as a third matrix. There is empirical evidence to suggest a positive relationship between temporal and craniometric distance within certain archaeological sites [47], but the exact parameters of an isolation-by-space and time model are currently unclear. Therefore, we follow the methods described by Pinhasi & von Cramon-Taubadel [3] in applying a model of isolation-by-geographical distance, with a correction for temporal autocorrelation, via partial Mantel tests. Mantel tests were used because matrix elements cannot be considered independent and therefore, significance was assessed via a randomization test (9999 permutations). Bonferroni correction was applied in all cases, yielding a critical α-value of 0.017 [27,31]. All Mantel tests were performed in Passage 1.1, freely available online (www.passagesoftware.net).
Dow–Cheverud tests were employed in order to ascertain whether any of the five model matrices were significantly different in their congruence with the craniometric distance matrix [50]. The null hypothesis is that the correlations between two dependent matrices (B and C) and one independent matrix (A) are equal. Therefore, if the hypothesis is rejected at p ≤ 0.05, one of the dependent matrices is significantly more strongly correlated with the independent matrix than the other. In order to control for temporal distance, each of the five geographical model matrices and the craniometric distance matrix were first regressed onto the temporal distance matrix. Thereafter, the residuals were employed as input data for the Dow–Cheverud tests. All comparisons were performed in R using a code written by Mark Grabowski and Charles Roseman. Dow–Cheverud comparisons were only performed between those alternative models that were significantly correlated with the craniometric data based on the partial Mantel tests. We add the caveat that all significance values are considered minimum values given the inherent error in the estimation of biological matrices [51] and given that Dow–Cheverud tests may be susceptible to type I errors when the data are spatially autocorrelated [51,52].
3. Results
(a). Population affinity patterns
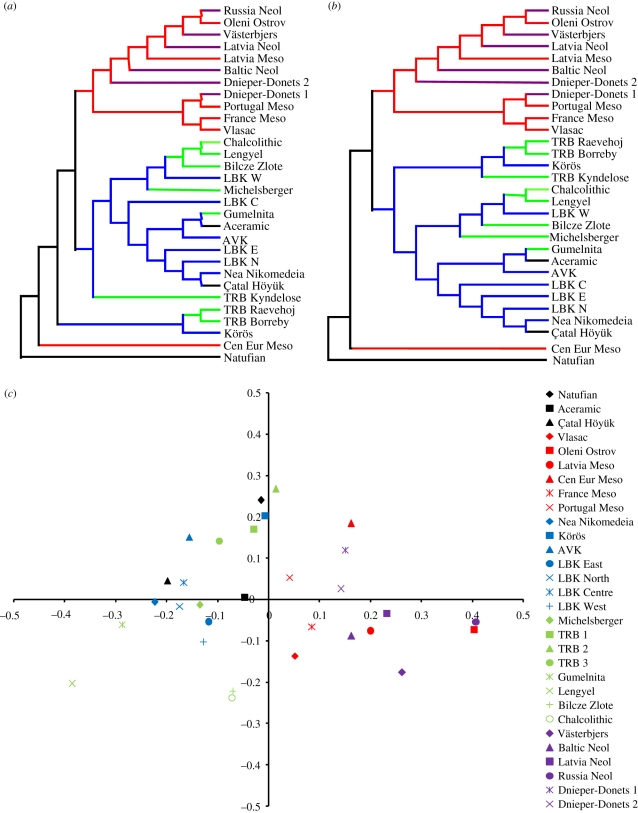
Craniometric distances between OTUs are illustrated in figure 3 in the form of two phenetic trees and a plot of the first two principal coordinates. In all cases, there is a clear distinction between Early/Late true Neolithic OTUs (blue and green) and Mesolithic/Late Forest Neolithic (red and purple). Moreover, Early Neolithic populations from the Near East (black) consistently cluster with true Neolithic populations from across Europe.
Figure 3.
Craniometric affinities among 30 OTUs. Unrooted (a) neighbour-joining and (b) minimum evolution trees of craniometric distances between OTUs. (c) Plot of the first two principal coordinates of craniometric distance. PCO1 (x-axis) explains 32.2% of total variance, and PCO2 (y-axis) explains 16.2% variance.
(b). Correlation of craniometric distance and alternative models
The results of the partial Mantel tests, controlling for temporal distance, show that craniometric distances do not fit the null hypothesis of pure cultural diffusion (table 1), further supporting the demic diffusion model for the initial transition to agriculture. Model 1 was not significantly correlated with the craniometric data and Model 2 was significantly correlated with craniometric distance, but only when demic diffusion was modelled as being relatively strong (1000 and 1500 km). Models 3 and 4, which include gene flow barriers between Mesolithic and Neolithic lineages, were also significantly correlated with craniometric distance. Dow–Cheverud tests were used to test whether Model 4 (highest Mantel test r-value) was also significantly more likely to explain the craniometric data than Models 2 and 3 (weaker r-values). Dow–Cheverud tests found no significant difference between Models 2 and 3, but found that Model 4 explained the craniometric pattern significantly better than Models 2 and 3.
Table 1.
Results of the partial Mantel and Dow–Cheverud tests. (Partial Mantel test results are Bonferroni corrected (α ≤ 0.017) r-values (p-values in parentheses). Dow–Cheverud results are p1Z scores (p-values in parentheses). All significant results are shown in bold.)
| Partial Mantel tests | Null Model (pure cultural diffusion) | 0.146 (0.149) | ||
| 500 km | 1000 km | 1500 km | ||
| Model 1 | 0.176 (0.077) | 0.197 (0.076) | 0.208 (0.052) | |
| Model 2 | 0.206 (0.046) | 0.255 (0.011) | 0.286 (0.007) | |
| Model 3 | 0.249 (0.013) | 0.275 (0.008) | 0.303 (0.003) | |
| Model 4 | 0.257 (0.016) | 0.297 (0.002) | 0.322 (0.001) | |
| Dow–Cheverud tests | Model 2 versus Model 3 | 0.129 (0.103) | 0.110 (0.114) | |
| Model 2 versus Model 4 | 0.260 (0.001) | 0.238 (0.004) | ||
| Model 3 versus Model 4 | 0.194 (0.019) | 0.181 (0.026) | 0.172 (0.022) |
4. Discussion
The results clearly show that the craniometric affinity patterns are best explained by a model that involves a barrier to gene flow between farming and hunter–gatherer populations. It must be emphasized that the impediment to gene flow was modelled as being relatively weak and, therefore, it is likely that the strength of the gene flow barrier between the farming and non-farming lineages was underestimated rather than overestimated. Our models never exclude gene flow between any contemporaneous populations regardless of their archaeological labels. Instead, what the results show is that there was relatively less gene flow between contemporaneous populations of the two lineages, therefore creating the observed craniometric patterns (figure 3). Moreover, the fact that outlying hunter–gatherer populations adopted some cultural elements from contemporaneous farming communities suggests some cultural (and possibly biological) interaction. However, the two dichotomous lineages, clearly identifiable in the craniometric data, show that the transition to agriculture in remote regions of Europe involved a complex mosaic of demic diffusion, cultural diffusion and changes in admixture patterns.
The results presented here illustrate that the use of two opposing mutually exclusive models of demic versus cultural diffusion is an oversimplification of what was a more complicated demographic transition in Europe. They also make clear that the transition to farming in Europe did not involve the complete replacement of indigenous hunter–gatherer populations despite significant gene flow from the Near East. The use of a mosaic model of biological and cultural change also helps reconcile the conflicting results often obtained from analyses of modern DNA (e.g. [5–12]) and from archaeological studies (e.g. [4,26]), whereby studies suggest either demic or cultural diffusion as being the principal basis for change. Further refining this mosaic model and understanding its underlying environmental, ecological and social causes present the most important future challenge.
Acknowledgements
We are very grateful to John Relethford for kindly making available his RMET software and to Charles Roseman and Mark Grabowski for providing their R code for the Dow–Cheverud test. We thank Lia Betti, Stephen Lycett, Russell Gray and two anonymous reviewers for constructive comments that much improved our manuscript. We also wish to acknowledge Winfried Heneke for kindly providing access to craniometric data. This research was supported by the European Research Council Starting Grant (ERC-2010-StG 263441).
References
- 1.Whittle A., Cummings V. (eds) 2007. Going over: the Mesolithic–Neolithic transition in north-west Europe. Oxford, UK: Oxford University Press [Google Scholar]
- 2.Ammerman A. J., Cavalli-Sforza L. L. 1984. The Neolithic transition and the genetics of populations in Europe. Princeton, NJ: Princeton University Press [Google Scholar]
- 3.Pinhasi R., von Cramon-Taubadel N. 2009. Craniometric data supports demic diffusion model for the spread of agriculture into Europe. PLoS ONE 4, e6747. 10.1371/journal.pone.0006747 (doi:10.1371/journal.pone.0006747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey G., Spikins P. (eds) 2008. Mesolithic Europe. Cambridge, UK: Cambridge University Press [Google Scholar]
- 5.Richards M., et al. 2000. Tracing European founder lineages in the near eastern mtDNA pool. Am. J. Hum. Genet. 67, 1251–1276 [PMC free article] [PubMed] [Google Scholar]
- 6.Torroni A., et al. 2001. A signal, from human mtDNA of postglacial recolonisation in Europe. Am. J. Hum. Genet. 69, 844–852 10.1086/323485 (doi:10.1086/323485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikhi L., Nichols R. A., Barbujani G., Beaumont M. A. 2002. Y genetic data support the Neolithic demic diffusion model. Proc. Natl Acad. Sci. USA 99, 11 008–11 013 10.1073/pnas.162158799 (doi:10.1073/pnas.162158799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells R. S., et al. 2007. The Eurasian Heartland: a continental perspective on Y-chromosome diversity. Proc. Natl Acad. Sci. USA 98, 10 244–10 249 10.1073/pnas.171305098 (doi:10.1073/pnas.171305098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupanloup I., Bertorelle G., Chikhi L., Barbujani G. 2004. Estimating the impact of prehistoric admixture on the genome of Europeans. Mol. Biol. Evol. 21, 1361–1372 10.1093/molbev/msh135 (doi:10.1093/molbev/msh135) [DOI] [PubMed] [Google Scholar]
- 10.Semino O., et al. 2004. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 74, 1023–1034 10.1086/386295 (doi:10.1086/386295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belle E. M., Landry P. A., Barbujani G. 2006. Origins and evolution of the Europeans' genome: evidence from multiple microsatellite loci. Proc. R. Soc. B 273, 1595–1602 10.1098/rspb.2006.3494 (doi:10.1098/rspb.2006.3494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itan Y., Powell A., Beaumont M. A., Burger J., Thomas M. G. 2009. The origins of lactase persistence in Europe. PLoS Comp. Biol. 5, e1000491. 10.1371/journal.pcbi.1000491 (doi:10.1371/journal.pcbi.1000491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haak W., et al. 2007. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science 310, 1016–1018 10.1126/science.1118725 (doi:10.1126/science.1118725) [DOI] [PubMed] [Google Scholar]
- 14.Sampietro M. L., Lao O., Caramelli D., Lari M., Pou R., Marti M., Bertranpetit J., Lalueza-Fox C. 2007. Palaeogenetic evidence supports a duel model of Neolithic spreading into Europe. Proc. R. Soc. B 274, 2161–2167 10.1098/rspb.2007.0465 (doi:10.1098/rspb.2007.0465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bramanti B., et al. 2009. Genetic discontinuity between local hunter–gatherers and Central Europe's first farmers. Science 326, 137–140 10.1126/science.1176869 (doi:10.1126/science.1176869) [DOI] [PubMed] [Google Scholar]
- 16.Helgason A., et al. 2009. Sequences from first settlers reveal rapid evolution in Icelandic mtDNA pool. PLoS Genetics 5, e1000343. 10.1371/journal.pgen.1000343 (doi:10.1371/journal.pgen.1000343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malmström H., et al. 2009. Ancient DNA reveals lack of continuity between Neolithic hunter–gatherers and contemporary Scandinavians. Curr. Biol. 19, 1758–1762 10.1016/j.cub.2009.09.017 (doi:10.1016/j.cub.2009.09.017) [DOI] [PubMed] [Google Scholar]
- 18.Ghirotto S., Mona S., Benazzo A., Paparazzo F., Caramelli D., Barbujani G. 2010. Inferring genealogical processes from patterns of Bronze-age and modern DNA variation in Sardinia. Mol. Biol. Evol. 27, 875–886 10.1093/molbev/msp292 (doi:10.1093/molbev/msp292) [DOI] [PubMed] [Google Scholar]
- 19.Haak W., et al. 2010. Ancient DNA from European early Neolithic farmers reveals their near eastern affinities. PLoS Biol. 8, e1000536. 10.1371/journal.pbio.1000536 (doi:10.1371/journal.pbio.1000536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-Yosef O. 2004. East to west: agricultural origins and dispersal into Europe. Curr. Anthropol. 45, S1–S3 10.1086/423970 (doi:10.1086/423970) [DOI] [Google Scholar]
- 21.Pinhasi R., Pluciennik M. 2004. A regional biological approach to the spread of farming in Europe. Curr. Anthropol. 45, S59–S82 10.1086/422085 (doi:10.1086/422085) [DOI] [Google Scholar]
- 22.Zvelebil M. 1986. Mesolithic societies and the transition to farming: problems of time, scale and organization. In Hunters in transition: Mesolithic societies of temperate Eurasia and their transition to farming (eds Zvelebil M., Dennell R., Domanka L.), pp. 167–188 Cambridge, UK: Cambridge University Press [Google Scholar]
- 23.Zvelebil M., Dolukhanov P. 1991. Transition to farming in Eastern and Northern Europe. J. Wor. Prehist. 5, 233–278 10.1007/BF00974991 (doi:10.1007/BF00974991) [DOI] [Google Scholar]
- 24.Price T. D. 1996. The first farmers of southern Scandinavia. In The origins and spread of agriculture and pastoralism in Eurasia (ed. Harris D. R.), pp. 346–362 London, UK: UCL Press [Google Scholar]
- 25.Thomas J. 1996. The cultural context of the first use of domesticates in continental Central and Northwestern Europe. In The origins and spread of agriculture and pastoralism in Eurasia (ed. Harris D. R.), pp. 310–322 London, UK: UCL Press [Google Scholar]
- 26.Zvelebil M. 1996. The agricultural frontier and the transition to farming in the circum-Baltic area. In The origins and spread of agriculture and pastoralism in Eurasia (ed. Harris D. R.), pp. 323–345 London, UK: UCL Press [Google Scholar]
- 27.Roseman C. C. 2004. Detecting interregionally diversifying natural selection on modern human cranial form by using matched molecular and morphometric data. Proc. Natl Acad. Sci. USA 101, 12 824–12 829 10.1073/pnas.0402637101 (doi:10.1073/pnas.0402637101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Relethford J. H. 2004. Global patterns of isolation by distance based on genetic and morphological data. Hum. Biol. 76, 499–513 10.1353/hub.2004.0060 (doi:10.1353/hub.2004.0060) [DOI] [PubMed] [Google Scholar]
- 29.Harvati K., Weaver T. D. 2006. Human cranial anatomy and the differential preservation of population history and climate signatures. Anat. Rec. 288A, 1225–1233 10.1002/ar.a.20395 (doi:10.1002/ar.a.20395) [DOI] [PubMed] [Google Scholar]
- 30.Manica A., Amos W., Balloux F., Hanihara T. 2007. The effect of ancient population bottlenecks on human phenotypic variation. Nature 448, 346–349 10.1038/nature05951 (doi:10.1038/nature05951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Cramon-Taubadel N. 2009. Congruence of individual cranial bone morphology and neutral molecular affinity patterns in modern humans. Am. J. Phys. Anthropol. 140, 205–215 10.1002/ajpa.21041 (doi:10.1002/ajpa.21041) [DOI] [PubMed] [Google Scholar]
- 32.Betti L., Balloux F., Amos W., Hanihara T., Manica A. 2009. Distance from Africa, not climate, explains within-population phenotypic diversity in humans. Proc. R. Soc. B 276, 809–814 10.1098/rspb.2008.1563 (doi:10.1098/rspb.2008.1563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Cramon-Taubadel N., Weaver T. D. 2009. Insights from a quantitative genetic approach to human morphological evolution. Evol. Anthropol. 18, 237–240 10.1002/evan.20233 (doi:10.1002/evan.20233) [DOI] [Google Scholar]
- 34.Scherer A. K. 2007. Population structure of the classic period Maya. Am. J. Phys. Anthropol. 132, 367–380 10.1002/ajpa.20535 (doi:10.1002/ajpa.20535) [DOI] [PubMed] [Google Scholar]
- 35.Brauer G. 1988. Osteometrie. In Anthropologie Handbuch der Vergleichenden Biologie des Menschen Band, 1st edn (ed. Knussmann R.), pp. 160–232 Stuttgart, Germany: Gustav Fischer Verlag [Google Scholar]
- 36.Buikstra J. E., Ubelaker D. 1994. Standards for data collection from human skeletal remains. Arkansas Archaeological Survey Research Series no. 44. Fayetteville, AR: Arkansas Archaeological Survey Research [Google Scholar]
- 37.Martin R., Saller K. 1957. Lehrbuch der Anthropologie in Systematischer Darstellung, 3rd edn. Stuttgart, Germany: Gustav Fischer Verlag [Google Scholar]
- 38.Falsetti A. B., Jungers W. L., Cole I. T. M. 1993. Morphometrics of the Callitrichid forelimb: a case study in size and shape. Int. J. Primatol. 14, 551–572 10.1007/BF02215447 (doi:10.1007/BF02215447) [DOI] [Google Scholar]
- 39.Jungers W. L., Falsetti A. B., Wall C. E. 1995. Shape, relative size and size-adjustments in morphometrics. Yrbk Phys. Anthropol. 38, 137–161 10.1002/ajpa.1330380608 (doi:10.1002/ajpa.1330380608) [DOI] [Google Scholar]
- 40.Relethford J. H., Blangero J. 1990. Detection of differential gene flow from patterns of quantitative variation. Hum. Biol. 62, 5–25 [PubMed] [Google Scholar]
- 41.Rzhetsky A., Nei M. 1993. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol. Biol. Evol. 10, 1073–1095 [DOI] [PubMed] [Google Scholar]
- 42.Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 43.Kumar S., Tamura K., Nei M. 2004. MEGA3: integrated software for molecular and evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5, 150–163 10.1093/bib/5.2.150 (doi:10.1093/bib/5.2.150) [DOI] [PubMed] [Google Scholar]
- 44.Page R. D. M. 1996. Treeview: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 45.Waddle D. M. 1994. Matrix correlation tests support a single origin for modern humans. Nature 368, 452–454 10.1038/368452a0 (doi:10.1038/368452a0) [DOI] [PubMed] [Google Scholar]
- 46.Konigsberg L. W. 1990. Analysis of prehistoric biological variation under a model of isolation by geographic and temporal distance. Hum. Biol. 62, 49–70 [PubMed] [Google Scholar]
- 47.Konigsberg L. W. 1990. Temporal aspects of biological distance: serial correlation and trend in a prehistoric skeletal lineage. Am. J. Phys. Anthropol. 82, 45–52 10.1002/ajpa.1330820106 (doi:10.1002/ajpa.1330820106) [DOI] [PubMed] [Google Scholar]
- 48.Sinnott R. W. 1984. Virtues of the haversine. Sky Telesc. 68, 159 [Google Scholar]
- 49.Mantel N. A. 1967. The detection of disease clustering and a generalized regression approach. Can. Res. 27, 209–220 [PubMed] [Google Scholar]
- 50.Dow M. M., Cheverud J. M. 1985. Comparison of distance matrices in studies of population structure and genetic microdifferentiation: quadratic assignment. Am. J. Phys. Anthropol. 68, 367–373 10.1002/ajpa.1330680307 (doi:10.1002/ajpa.1330680307) [DOI] [PubMed] [Google Scholar]
- 51.Konigsberg L. W. 1997. Comments on matrix permutation tests in the evaluation of competing models for modern human origins. J. Hum. Evol. 32, 479–488 10.1006/jhev.1996.0125 (doi:10.1006/jhev.1996.0125) [DOI] [PubMed] [Google Scholar]
- 52.Oden N. L. 1992. Spatial autocorrelation invalidates the Dow–Cheverud test. Am. J. Phys. Anthropol. 89, 257–264 10.1002/ajpa.1330890209 (doi:10.1002/ajpa.1330890209) [DOI] [Google Scholar]