Abstract
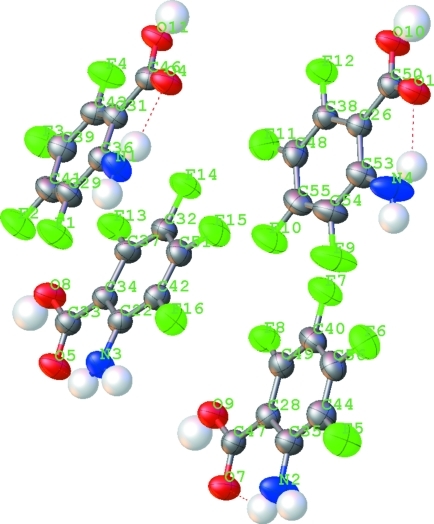
The asymmetric unit of the title compound, C7H3F4NO2, obtained as an intermediate in the synthesis of a coupling reagent, contains four independent and conformationally similar molecules. The amine H atoms form both intramolecular and intermolecular N—H⋯Ocarboxyl hydrogen bonds which, together with intermolecular O—H⋯Ocarboxyl hydrogen bonds and N—H⋯F associations form ribbon structures along the a axis.
Related literature
The title compound was obtained as one of the intermediates in the synthesis of a coupling reagent (Xu et al., 2008 ▶; Liao et al., 2007 ▶), using the Hofmann rearrangement (Perumal & Muthialu, 2004 ▶) with 2-carboxyl-3,4,5,6-tetrafluorobenzamide (Cai et al., 1992 ▶).
Experimental
Crystal data
C7H3F4NO2
M r = 209.10
Triclinic,

a = 11.0367 (11) Å
b = 11.3664 (11) Å
c = 12.5702 (12) Å
α = 80.378 (8)°
β = 79.764 (8)°
γ = 82.011 (8)°
V = 1520.2 (3) Å3
Z = 8
Cu Kα radiation
μ = 1.79 mm−1
T = 295 K
0.50 × 0.30 × 0.15 mm
Data collection
Oxford Diffraction Xcalibur Sapphire3 Gemini ultra CCD diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010 ▶) T min = 0.392, T max = 1.000
8708 measured reflections
4791 independent reflections
3521 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.057
wR(F 2) = 0.177
S = 1.05
4791 reflections
533 parameters
16 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.43 e Å−3
Δρmin = −0.24 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2010 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: OLEX2.
Supplementary Material
Crystal structure: contains datablock(s) global. DOI: 10.1107/S1600536811023087/zs2117sup1.cif
Supplementary material file. DOI: 10.1107/S1600536811023087/zs2117globalsup2.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1B⋯O4 | 0.84 (3) | 2.04 (4) | 2.637 (4) | 128 (3) |
| N1—H1B⋯F5i | 0.84 (3) | 2.40 (3) | 3.154 (4) | 150 (3) |
| N2—H2A⋯O10ii | 0.83 (3) | 2.58 (3) | 3.363 (5) | 158 (3) |
| N2—H2B⋯O7 | 0.85 (3) | 2.03 (4) | 2.643 (5) | 129 (3) |
| N3—H3A⋯O11iii | 0.85 (3) | 2.50 (3) | 3.337 (4) | 167 (3) |
| N3—H3B⋯O5 | 0.85 (3) | 2.04 (4) | 2.654 (4) | 129 (3) |
| N4—H4B⋯O1 | 0.86 (3) | 2.01 (3) | 2.648 (4) | 130 (3) |
| N4—H4B⋯F16iv | 0.86 (3) | 2.48 (3) | 3.191 (5) | 140 (3) |
| O9—H9⋯O5v | 0.82 | 1.84 | 2.660 (3) | 175 |
| O10—H10⋯O4vi | 0.82 | 1.86 | 2.675 (4) | 178 |
| O11—H11⋯O1vi | 0.82 | 1.82 | 2.643 (3) | 177 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  .
.
Acknowledgments
This work was supported by grants from the National High Technology Development Project (863 Project; Nos. 2006 A A09Z408 GDSFC 06025194, 2005 A30503001, and 2006Z3E4041) and the National Natural Science Fund (No. 20772048)
supplementary crystallographic information
Comment
The title compound C7H3F4NO2 (I) was obtained as one of the intermediates in the synthesis of a coupling reagent (Xu et al., 2008, Liao et al., 2007), using the Hofmann rearrangement (Perumal & Muthialu, 2004) with 2-carboxyl-3,4,5,6-tetrafluorobenzamide (Cai et al., 1992). In the structure of (I), the asymmetric unit contains four independent and conformationally similar molecules (Fig. 1). The molecules associate through carboxylic acid O—H···Ocarboxyl hydrogen bonds (Table 1) while the amine H atoms form both intramolecular N—H···Ocarboxyl hydrogen bonds as well as intermolecular N—H···F associations give one-dimensional ribbon structures. Also present in the structure are short intermolecular F···F contacts [minimum, 2.825 (3) Å].
Experimental
To a stirred solution of 39.2 g of KOH in 356 mL of distilled water, 11.3 g of bromine was added and after 30 min, 70 mmol of 2-carboxy-3,4,5,6-tetrafluorobenzamide was added. After allowing the reaction to proceed for 30 min at 293 K, the mixture was heated to 363K-368K and maintained at that temperature for 4 h, after which the mixture was cooled to room temperature and allowed to stand for 48 h. To the mixture was then added 100 mL of water, the pH adjusted to 1 at ice-water temperature, stirred and filtered, giving a yellow solid (16.2 g, yield 94%). Pale yellow crystals of (I) suitable for X-ray analysis grew over a period of a week from a solution of the solid in methanol at room temperature.
Refinement
The carboxylic acid H atoms were positioned geometrically and were included in the refinement in the riding-model approximation with O—H = 0.82 Å and Uiso(H) = 1.5Ueq(O) The amine H atoms were located in difference Fourier maps and the positional parameters were refined but with the displacement parameters riding with Uiso(H) = 1.5Ueq(N).
Figures
Fig. 1.
The molecular conformation of the four independent molecules in the asymmetric unit of the title compound, showing the atom numbering scheme. Intramolecular hydrogen bonds are shown as dashed lines. Displacement ellipsoids are drawn at the 50% probability level.
Crystal data
| C7H3F4NO2 | Z = 8 |
| Mr = 209.10 | F(000) = 832 |
| Triclinic, P1 | Dx = 1.827 Mg m−3 |
| a = 11.0367 (11) Å | Cu Kα radiation, λ = 1.5418 Å |
| b = 11.3664 (11) Å | Cell parameters from 4167 reflections |
| c = 12.5702 (12) Å | θ = 3.6–62.7° |
| α = 80.378 (8)° | µ = 1.79 mm−1 |
| β = 79.764 (8)° | T = 295 K |
| γ = 82.011 (8)° | Plate, pale yellow |
| V = 1520.2 (3) Å3 | 0.50 × 0.30 × 0.15 mm |
Data collection
| Oxford Diffraction Xcalibur Sapphire3 Gemini ultra CCD diffractometer | 4791 independent reflections |
| Radiation source: Enhance Ultra (Cu) X-ray Source | 3521 reflections with I > 2σ(I) |
| mirror | Rint = 0.031 |
| Detector resolution: 16.0288 pixels mm-1 | θmax = 62.8°, θmin = 3.6° |
| ω scans | h = −12→12 |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010) | k = −12→13 |
| Tmin = 0.392, Tmax = 1.000 | l = −14→14 |
| 8708 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.057 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.177 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0935P)2 + 0.6844P] where P = (Fo2 + 2Fc2)/3 |
| 4791 reflections | (Δ/σ)max < 0.001 |
| 533 parameters | Δρmax = 0.43 e Å−3 |
| 16 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F16 | 0.18790 (19) | 0.2980 (2) | 0.63873 (16) | 0.0672 (6) | |
| F13 | −0.0401 (2) | 0.42992 (19) | 0.28267 (16) | 0.0682 (6) | |
| F14 | 0.1447 (2) | 0.54588 (19) | 0.30496 (17) | 0.0701 (6) | |
| O8 | −0.1849 (2) | 0.2707 (2) | 0.3563 (2) | 0.0632 (7) | |
| H8 | −0.2349 | 0.2248 | 0.3531 | 0.095* | |
| F15 | 0.2604 (2) | 0.4812 (2) | 0.48248 (19) | 0.0747 (6) | |
| O5 | −0.1389 (2) | 0.1291 (2) | 0.49241 (19) | 0.0622 (7) | |
| C22 | 0.0365 (3) | 0.2621 (3) | 0.5411 (2) | 0.0441 (7) | |
| N3 | 0.0065 (3) | 0.1737 (3) | 0.6245 (2) | 0.0581 (8) | |
| C32 | 0.1097 (3) | 0.4535 (3) | 0.3810 (3) | 0.0502 (8) | |
| C33 | −0.1201 (3) | 0.2250 (3) | 0.4350 (2) | 0.0470 (7) | |
| C34 | −0.0239 (3) | 0.2952 (3) | 0.4488 (2) | 0.0443 (7) | |
| C37 | 0.0151 (3) | 0.3923 (3) | 0.3714 (2) | 0.0456 (7) | |
| C42 | 0.1311 (3) | 0.3284 (3) | 0.5489 (3) | 0.0491 (8) | |
| C51 | 0.1684 (3) | 0.4208 (3) | 0.4715 (3) | 0.0524 (8) | |
| F8 | 0.3963 (2) | 0.21194 (19) | 0.48315 (16) | 0.0667 (6) | |
| O9 | 0.2802 (2) | 0.0302 (2) | 0.5375 (2) | 0.0615 (6) | |
| H9 | 0.2341 | −0.0183 | 0.5317 | 0.092* | |
| O7 | 0.3410 (2) | −0.1190 (2) | 0.6616 (2) | 0.0627 (7) | |
| F5 | 0.6708 (2) | 0.0512 (2) | 0.8060 (2) | 0.0892 (8) | |
| F6 | 0.6953 (2) | 0.2663 (2) | 0.6819 (2) | 0.0787 (7) | |
| F7 | 0.5568 (2) | 0.3471 (2) | 0.5210 (2) | 0.0827 (7) | |
| C28 | 0.4414 (3) | 0.0572 (3) | 0.6287 (2) | 0.0462 (7) | |
| C35 | 0.5150 (3) | 0.0154 (3) | 0.7126 (3) | 0.0531 (8) | |
| C40 | 0.5419 (3) | 0.2401 (3) | 0.5830 (3) | 0.0577 (9) | |
| C44 | 0.5992 (3) | 0.0896 (3) | 0.7262 (3) | 0.0601 (9) | |
| C47 | 0.3507 (3) | −0.0174 (3) | 0.6118 (2) | 0.0469 (7) | |
| C49 | 0.4586 (3) | 0.1696 (3) | 0.5659 (3) | 0.0498 (8) | |
| N2 | 0.5075 (4) | −0.0899 (3) | 0.7795 (3) | 0.0833 (12) | |
| C56 | 0.6129 (3) | 0.1984 (3) | 0.6649 (3) | 0.0583 (9) | |
| F4 | 0.0302 (2) | 0.94676 (19) | 0.75391 (15) | 0.0700 (6) | |
| O11 | 0.1722 (2) | 1.0628 (2) | 0.82373 (17) | 0.0570 (6) | |
| H11 | 0.2109 | 1.1185 | 0.8268 | 0.085* | |
| O4 | 0.2335 (2) | 0.9962 (2) | 0.98460 (19) | 0.0594 (6) | |
| F3 | −0.1220 (2) | 0.7811 (2) | 0.78819 (17) | 0.0745 (6) | |
| F2 | −0.1394 (2) | 0.6232 (2) | 0.97776 (19) | 0.0815 (7) | |
| F1 | −0.0109 (2) | 0.6443 (2) | 1.13507 (17) | 0.0810 (7) | |
| N1 | 0.1387 (3) | 0.8133 (3) | 1.1147 (2) | 0.0620 (8) | |
| C29 | 0.0014 (3) | 0.7205 (3) | 1.0410 (3) | 0.0543 (8) | |
| C31 | 0.0926 (3) | 0.8903 (3) | 0.9301 (2) | 0.0432 (7) | |
| C36 | 0.0799 (3) | 0.8091 (3) | 1.0291 (2) | 0.0471 (7) | |
| C39 | −0.0549 (3) | 0.7890 (3) | 0.8658 (3) | 0.0544 (8) | |
| C41 | −0.0645 (3) | 0.7095 (3) | 0.9618 (3) | 0.0565 (9) | |
| C43 | 0.0231 (3) | 0.8750 (3) | 0.8509 (2) | 0.0473 (7) | |
| C46 | 0.1720 (3) | 0.9855 (3) | 0.9152 (2) | 0.0464 (7) | |
| F12 | 0.4204 (2) | 0.75684 (19) | −0.01077 (18) | 0.0705 (6) | |
| F11 | 0.2839 (2) | 0.5761 (2) | 0.0206 (2) | 0.0766 (7) | |
| O10 | 0.6074 (3) | 0.8442 (2) | 0.0225 (2) | 0.0687 (7) | |
| H10 | 0.6552 | 0.8943 | 0.0195 | 0.103* | |
| O1 | 0.6965 (2) | 0.7627 (2) | 0.1668 (2) | 0.0632 (7) | |
| C26 | 0.5475 (3) | 0.6595 (3) | 0.1206 (2) | 0.0451 (7) | |
| F10 | 0.3353 (2) | 0.3778 (2) | 0.1668 (2) | 0.0847 (7) | |
| F9 | 0.5247 (3) | 0.3629 (2) | 0.2782 (2) | 0.1045 (10) | |
| C38 | 0.4484 (3) | 0.6618 (3) | 0.0631 (3) | 0.0487 (8) | |
| N4 | 0.6652 (4) | 0.5390 (3) | 0.2570 (3) | 0.0928 (14) | |
| C48 | 0.3770 (3) | 0.5711 (3) | 0.0787 (3) | 0.0544 (8) | |
| C50 | 0.6226 (3) | 0.7589 (3) | 0.1051 (3) | 0.0497 (8) | |
| C53 | 0.5726 (4) | 0.5559 (3) | 0.1963 (3) | 0.0582 (9) | |
| C54 | 0.4979 (4) | 0.4638 (3) | 0.2087 (3) | 0.0660 (10) | |
| C55 | 0.4029 (4) | 0.4695 (3) | 0.1527 (3) | 0.0597 (9) | |
| H3A | 0.059 (3) | 0.147 (3) | 0.667 (2) | 0.072* | |
| H3B | −0.039 (3) | 0.123 (3) | 0.615 (3) | 0.072* | |
| H2A | 0.551 (3) | −0.113 (3) | 0.828 (2) | 0.100* | |
| H2B | 0.463 (3) | −0.140 (2) | 0.768 (3) | 0.100* | |
| H1A | 0.129 (3) | 0.760 (3) | 1.1720 (19) | 0.074* | |
| H1B | 0.186 (3) | 0.866 (3) | 1.112 (3) | 0.074* | |
| H4A | 0.681 (3) | 0.476 (2) | 0.299 (3) | 0.107* | |
| H4B | 0.713 (3) | 0.595 (2) | 0.249 (3) | 0.107* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F16 | 0.0711 (13) | 0.0830 (14) | 0.0560 (11) | −0.0196 (11) | −0.0327 (10) | −0.0008 (10) |
| F13 | 0.0819 (14) | 0.0688 (13) | 0.0578 (11) | −0.0236 (11) | −0.0350 (10) | 0.0176 (10) |
| F14 | 0.0769 (14) | 0.0623 (13) | 0.0693 (13) | −0.0277 (11) | −0.0150 (11) | 0.0155 (10) |
| O8 | 0.0711 (16) | 0.0587 (15) | 0.0679 (15) | −0.0207 (12) | −0.0400 (13) | 0.0094 (12) |
| F15 | 0.0702 (14) | 0.0751 (14) | 0.0891 (15) | −0.0329 (11) | −0.0289 (11) | −0.0035 (12) |
| O5 | 0.0732 (16) | 0.0615 (15) | 0.0586 (14) | −0.0300 (13) | −0.0300 (12) | 0.0119 (12) |
| C22 | 0.0460 (17) | 0.0459 (17) | 0.0392 (15) | −0.0039 (14) | −0.0091 (13) | −0.0018 (13) |
| N3 | 0.0614 (18) | 0.0679 (19) | 0.0467 (15) | −0.0201 (15) | −0.0227 (13) | 0.0121 (14) |
| C32 | 0.0527 (19) | 0.0426 (17) | 0.0533 (18) | −0.0106 (15) | −0.0077 (15) | 0.0028 (15) |
| C33 | 0.0510 (18) | 0.0487 (18) | 0.0421 (16) | −0.0080 (15) | −0.0120 (14) | −0.0019 (14) |
| C34 | 0.0477 (17) | 0.0444 (17) | 0.0414 (15) | −0.0077 (14) | −0.0125 (13) | 0.0001 (13) |
| C37 | 0.0526 (18) | 0.0442 (17) | 0.0402 (16) | −0.0077 (14) | −0.0163 (13) | 0.0046 (13) |
| C42 | 0.0504 (18) | 0.0532 (19) | 0.0455 (16) | −0.0057 (15) | −0.0171 (14) | −0.0023 (15) |
| C51 | 0.0479 (18) | 0.055 (2) | 0.0574 (19) | −0.0131 (15) | −0.0134 (15) | −0.0069 (16) |
| F8 | 0.0722 (13) | 0.0644 (13) | 0.0647 (12) | −0.0157 (10) | −0.0253 (10) | 0.0096 (10) |
| O9 | 0.0639 (15) | 0.0617 (15) | 0.0664 (15) | −0.0199 (12) | −0.0318 (12) | 0.0028 (12) |
| O7 | 0.0704 (16) | 0.0519 (14) | 0.0726 (15) | −0.0180 (12) | −0.0341 (13) | 0.0049 (12) |
| F5 | 0.0981 (18) | 0.0973 (18) | 0.0898 (16) | −0.0335 (14) | −0.0588 (14) | 0.0020 (14) |
| F6 | 0.0734 (14) | 0.0865 (16) | 0.0894 (15) | −0.0374 (12) | −0.0187 (12) | −0.0206 (13) |
| F7 | 0.0925 (17) | 0.0697 (14) | 0.0882 (16) | −0.0367 (13) | −0.0184 (13) | 0.0091 (12) |
| C28 | 0.0464 (17) | 0.0485 (18) | 0.0459 (17) | −0.0093 (14) | −0.0086 (14) | −0.0083 (14) |
| C35 | 0.059 (2) | 0.054 (2) | 0.0506 (18) | −0.0128 (16) | −0.0177 (15) | −0.0036 (15) |
| C40 | 0.062 (2) | 0.052 (2) | 0.060 (2) | −0.0190 (17) | −0.0034 (17) | −0.0053 (16) |
| C44 | 0.064 (2) | 0.068 (2) | 0.056 (2) | −0.0163 (19) | −0.0233 (17) | −0.0101 (18) |
| C47 | 0.0461 (17) | 0.0507 (19) | 0.0457 (16) | −0.0047 (14) | −0.0126 (13) | −0.0069 (15) |
| C49 | 0.0493 (18) | 0.056 (2) | 0.0443 (16) | −0.0094 (15) | −0.0093 (14) | −0.0033 (15) |
| N2 | 0.108 (3) | 0.072 (2) | 0.084 (2) | −0.037 (2) | −0.061 (2) | 0.0190 (19) |
| C56 | 0.055 (2) | 0.066 (2) | 0.060 (2) | −0.0208 (17) | −0.0073 (16) | −0.0166 (18) |
| F4 | 0.1041 (17) | 0.0657 (13) | 0.0480 (11) | −0.0320 (12) | −0.0316 (11) | 0.0086 (9) |
| O11 | 0.0755 (16) | 0.0548 (14) | 0.0449 (12) | −0.0271 (12) | −0.0167 (11) | 0.0045 (10) |
| O4 | 0.0715 (16) | 0.0568 (14) | 0.0564 (13) | −0.0259 (12) | −0.0274 (12) | 0.0074 (11) |
| F3 | 0.0953 (16) | 0.0776 (14) | 0.0655 (13) | −0.0313 (12) | −0.0364 (12) | −0.0099 (11) |
| F2 | 0.0936 (17) | 0.0807 (15) | 0.0808 (15) | −0.0517 (13) | −0.0238 (12) | 0.0036 (12) |
| F1 | 0.0971 (17) | 0.0848 (16) | 0.0630 (13) | −0.0440 (13) | −0.0275 (12) | 0.0279 (12) |
| N1 | 0.069 (2) | 0.077 (2) | 0.0442 (15) | −0.0264 (16) | −0.0210 (14) | 0.0075 (15) |
| C29 | 0.059 (2) | 0.058 (2) | 0.0457 (17) | −0.0195 (17) | −0.0085 (15) | 0.0060 (15) |
| C31 | 0.0484 (17) | 0.0452 (17) | 0.0376 (15) | −0.0105 (14) | −0.0079 (13) | −0.0051 (13) |
| C36 | 0.0507 (18) | 0.0489 (18) | 0.0426 (16) | −0.0084 (15) | −0.0100 (13) | −0.0043 (14) |
| C39 | 0.063 (2) | 0.060 (2) | 0.0478 (18) | −0.0139 (17) | −0.0203 (15) | −0.0117 (16) |
| C41 | 0.061 (2) | 0.053 (2) | 0.059 (2) | −0.0229 (17) | −0.0120 (16) | −0.0037 (16) |
| C43 | 0.061 (2) | 0.0457 (17) | 0.0366 (15) | −0.0092 (15) | −0.0140 (14) | −0.0014 (13) |
| C46 | 0.0519 (18) | 0.0437 (17) | 0.0436 (16) | −0.0070 (14) | −0.0087 (14) | −0.0043 (14) |
| F12 | 0.0690 (13) | 0.0618 (13) | 0.0836 (14) | −0.0162 (10) | −0.0357 (11) | 0.0138 (11) |
| F11 | 0.0623 (13) | 0.0722 (14) | 0.1048 (17) | −0.0188 (11) | −0.0332 (12) | −0.0095 (13) |
| O10 | 0.0822 (18) | 0.0604 (15) | 0.0701 (16) | −0.0318 (13) | −0.0379 (14) | 0.0190 (13) |
| O1 | 0.0750 (16) | 0.0601 (15) | 0.0614 (14) | −0.0257 (13) | −0.0317 (13) | 0.0084 (12) |
| C26 | 0.0489 (17) | 0.0449 (17) | 0.0435 (16) | −0.0083 (14) | −0.0119 (13) | −0.0044 (14) |
| F10 | 0.0967 (18) | 0.0647 (14) | 0.1022 (17) | −0.0426 (13) | −0.0251 (14) | −0.0014 (13) |
| F9 | 0.155 (3) | 0.0682 (15) | 0.1023 (19) | −0.0487 (17) | −0.0663 (18) | 0.0320 (14) |
| C38 | 0.0496 (18) | 0.0476 (18) | 0.0487 (17) | −0.0077 (15) | −0.0109 (14) | −0.0010 (14) |
| N4 | 0.129 (3) | 0.061 (2) | 0.105 (3) | −0.040 (2) | −0.079 (3) | 0.028 (2) |
| C48 | 0.0472 (19) | 0.059 (2) | 0.062 (2) | −0.0116 (16) | −0.0123 (15) | −0.0159 (17) |
| C50 | 0.0506 (18) | 0.0513 (19) | 0.0488 (17) | −0.0112 (15) | −0.0113 (14) | −0.0038 (15) |
| C53 | 0.070 (2) | 0.056 (2) | 0.0538 (19) | −0.0184 (18) | −0.0208 (17) | −0.0015 (16) |
| C54 | 0.094 (3) | 0.049 (2) | 0.058 (2) | −0.023 (2) | −0.0221 (19) | 0.0061 (17) |
| C55 | 0.068 (2) | 0.051 (2) | 0.064 (2) | −0.0235 (17) | −0.0069 (18) | −0.0102 (17) |
Geometric parameters (Å, °)
| F16—C42 | 1.360 (3) | F4—C43 | 1.345 (3) |
| F13—C37 | 1.344 (3) | O11—H11 | 0.8200 |
| F14—C32 | 1.346 (4) | O11—C46 | 1.325 (4) |
| O8—H8 | 0.8200 | O4—C46 | 1.227 (4) |
| O8—C33 | 1.318 (4) | F3—C39 | 1.346 (4) |
| F15—C51 | 1.341 (4) | F2—C41 | 1.337 (4) |
| O5—C33 | 1.224 (4) | F1—C29 | 1.341 (4) |
| C22—N3 | 1.354 (4) | N1—C36 | 1.362 (4) |
| C22—C34 | 1.412 (4) | N1—H1A | 0.859 (17) |
| C22—C42 | 1.396 (5) | N1—H1B | 0.846 (17) |
| N3—H3A | 0.855 (17) | C29—C36 | 1.390 (5) |
| N3—H3B | 0.854 (17) | C29—C41 | 1.364 (5) |
| C32—C37 | 1.365 (5) | C31—C36 | 1.417 (4) |
| C32—C51 | 1.381 (5) | C31—C43 | 1.406 (4) |
| C33—C34 | 1.463 (4) | C31—C46 | 1.454 (4) |
| C34—C37 | 1.405 (4) | C39—C41 | 1.379 (5) |
| C42—C51 | 1.361 (5) | C39—C43 | 1.360 (5) |
| F8—C49 | 1.335 (4) | F12—C38 | 1.341 (4) |
| O9—H9 | 0.8200 | F11—C48 | 1.353 (4) |
| O9—C47 | 1.320 (4) | O10—H10 | 0.8200 |
| O7—C47 | 1.228 (4) | O10—C50 | 1.314 (4) |
| F5—C44 | 1.363 (4) | O1—C50 | 1.229 (4) |
| F6—C56 | 1.337 (4) | C26—C38 | 1.410 (5) |
| F7—C40 | 1.344 (4) | C26—C50 | 1.460 (4) |
| C28—C35 | 1.424 (4) | C26—C53 | 1.413 (5) |
| C28—C47 | 1.462 (4) | F10—C55 | 1.334 (4) |
| C28—C49 | 1.403 (5) | F9—C54 | 1.353 (4) |
| C35—C44 | 1.388 (5) | C38—C48 | 1.351 (5) |
| C35—N2 | 1.347 (5) | N4—C53 | 1.356 (5) |
| C40—C49 | 1.367 (5) | N4—H4A | 0.827 (17) |
| C40—C56 | 1.385 (5) | N4—H4B | 0.869 (17) |
| C44—C56 | 1.356 (5) | C48—C55 | 1.388 (5) |
| N2—H2A | 0.829 (17) | C53—C54 | 1.393 (5) |
| N2—H2B | 0.851 (17) | C54—C55 | 1.352 (5) |
| C33—O8—H8 | 109.5 | C46—O11—H11 | 109.5 |
| N3—C22—C34 | 124.9 (3) | C36—N1—H1A | 120 (2) |
| N3—C22—C42 | 117.6 (3) | C36—N1—H1B | 121 (2) |
| C42—C22—C34 | 117.4 (3) | H1A—N1—H1B | 119 (3) |
| C22—N3—H3A | 118 (2) | F1—C29—C36 | 118.3 (3) |
| C22—N3—H3B | 119 (2) | F1—C29—C41 | 119.1 (3) |
| H3A—N3—H3B | 115 (3) | C41—C29—C36 | 122.6 (3) |
| F14—C32—C37 | 121.1 (3) | C36—C31—C46 | 120.2 (3) |
| F14—C32—C51 | 119.9 (3) | C43—C31—C36 | 116.8 (3) |
| C37—C32—C51 | 119.0 (3) | C43—C31—C46 | 123.0 (3) |
| O8—C33—C34 | 116.0 (3) | N1—C36—C29 | 117.5 (3) |
| O5—C33—O8 | 121.8 (3) | N1—C36—C31 | 124.0 (3) |
| O5—C33—C34 | 122.2 (3) | C29—C36—C31 | 118.5 (3) |
| C22—C34—C33 | 119.3 (3) | F3—C39—C41 | 120.3 (3) |
| C37—C34—C22 | 117.9 (3) | F3—C39—C43 | 120.7 (3) |
| C37—C34—C33 | 122.7 (3) | C43—C39—C41 | 119.0 (3) |
| F13—C37—C32 | 116.2 (3) | F2—C41—C29 | 120.2 (3) |
| F13—C37—C34 | 121.0 (3) | F2—C41—C39 | 120.1 (3) |
| C32—C37—C34 | 122.8 (3) | C29—C41—C39 | 119.7 (3) |
| F16—C42—C22 | 117.5 (3) | F4—C43—C31 | 121.1 (3) |
| F16—C42—C51 | 119.2 (3) | F4—C43—C39 | 115.5 (3) |
| C51—C42—C22 | 123.3 (3) | C39—C43—C31 | 123.4 (3) |
| F15—C51—C32 | 119.5 (3) | O11—C46—C31 | 116.6 (3) |
| F15—C51—C42 | 121.0 (3) | O4—C46—O11 | 121.3 (3) |
| C42—C51—C32 | 119.5 (3) | O4—C46—C31 | 122.0 (3) |
| C47—O9—H9 | 109.5 | C50—O10—H10 | 109.5 |
| C35—C28—C47 | 119.2 (3) | C38—C26—C50 | 122.7 (3) |
| C49—C28—C35 | 118.2 (3) | C38—C26—C53 | 117.4 (3) |
| C49—C28—C47 | 122.6 (3) | C53—C26—C50 | 119.9 (3) |
| C44—C35—C28 | 117.2 (3) | F12—C38—C26 | 120.4 (3) |
| N2—C35—C28 | 124.5 (3) | F12—C38—C48 | 116.6 (3) |
| N2—C35—C44 | 118.3 (3) | C48—C38—C26 | 123.0 (3) |
| F7—C40—C49 | 121.2 (3) | C53—N4—H4A | 123 (2) |
| F7—C40—C56 | 120.2 (3) | C53—N4—H4B | 118 (2) |
| C49—C40—C56 | 118.6 (3) | H4A—N4—H4B | 118 (3) |
| F5—C44—C35 | 118.1 (3) | F11—C48—C55 | 119.7 (3) |
| C56—C44—F5 | 118.6 (3) | C38—C48—F11 | 121.1 (3) |
| C56—C44—C35 | 123.3 (3) | C38—C48—C55 | 119.2 (3) |
| O9—C47—C28 | 116.0 (3) | O10—C50—C26 | 116.9 (3) |
| O7—C47—O9 | 121.3 (3) | O1—C50—O10 | 121.6 (3) |
| O7—C47—C28 | 122.8 (3) | O1—C50—C26 | 121.5 (3) |
| F8—C49—C28 | 121.3 (3) | N4—C53—C26 | 124.7 (3) |
| F8—C49—C40 | 116.1 (3) | N4—C53—C54 | 117.5 (3) |
| C40—C49—C28 | 122.6 (3) | C54—C53—C26 | 117.8 (3) |
| C35—N2—H2A | 123 (2) | F9—C54—C53 | 117.9 (3) |
| C35—N2—H2B | 120 (2) | C55—C54—F9 | 118.9 (3) |
| H2A—N2—H2B | 117 (3) | C55—C54—C53 | 123.2 (3) |
| F6—C56—C40 | 119.2 (3) | F10—C55—C48 | 119.9 (3) |
| F6—C56—C44 | 120.7 (3) | F10—C55—C54 | 120.7 (3) |
| C44—C56—C40 | 120.2 (3) | C54—C55—C48 | 119.4 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···F1 | 0.86 (3) | 2.31 (3) | 2.655 (4) | 104 (2) |
| N1—H1B···O4 | 0.84 (3) | 2.04 (4) | 2.637 (4) | 128 (3) |
| N1—H1B···F5i | 0.84 (3) | 2.40 (3) | 3.154 (4) | 150 (3) |
| N2—H2A···F5 | 0.83 (3) | 2.39 (3) | 2.670 (5) | 101 (2) |
| N2—H2A···O10ii | 0.83 (3) | 2.58 (3) | 3.363 (5) | 158 (3) |
| N2—H2B···O7 | 0.85 (3) | 2.03 (4) | 2.643 (5) | 129 (3) |
| N3—H3A···F4iii | 0.85 (3) | 2.39 (3) | 2.816 (4) | 112 (3) |
| N3—H3A···F16 | 0.85 (3) | 2.32 (3) | 2.652 (4) | 104 (2) |
| N3—H3A···O11iii | 0.85 (3) | 2.50 (3) | 3.337 (4) | 167 (3) |
| N3—H3B···O5 | 0.85 (3) | 2.04 (4) | 2.654 (4) | 129 (3) |
| N4—H4A···F9 | 0.83 (3) | 2.36 (3) | 2.650 (5) | 101 (3) |
| N4—H4B···O1 | 0.86 (3) | 2.01 (3) | 2.648 (4) | 130 (3) |
| N4—H4B···F16iv | 0.86 (3) | 2.48 (3) | 3.191 (5) | 140 (3) |
| O9—H9···O5v | 0.82 | 1.84 | 2.660 (3) | 175 |
| O10—H10···O4vi | 0.82 | 1.86 | 2.675 (4) | 178 |
| O11—H11···O1vi | 0.82 | 1.82 | 2.643 (3) | 177 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) x, y−1, z+1; (iii) x, y−1, z; (iv) −x+1, −y+1, −z+1; (v) −x, −y, −z+1; (vi) −x+1, −y+2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZS2117).
References
- Cai, S.-X., Glenn, D. J. & Keana, J. F. W. (1992). J. Org. Chem. 57, 1299–1304.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Liao, X.-J., Xu, W.-J., Xu, S.-H. & Dong, F.-F. (2007). Acta Cryst. E63, o3313.
- Oxford Diffraction (2010). CrysAlis PRO Oxford Diffraction Ltd, Yarnton, England.
- Perumal, R. & Muthialu, S. (2004). Synth. Commun. 34, 1811–1818.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Xu, W.-J., Liao, X.-J., Xu, S.-H., Diao, J.-Z., Du, B., Zhou, X.-L. & Pan, S.-S. (2008). Org. Lett. 10, 4569–4572. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global. DOI: 10.1107/S1600536811023087/zs2117sup1.cif
Supplementary material file. DOI: 10.1107/S1600536811023087/zs2117globalsup2.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report