Abstract
The N-terminal region of human HOXA13 has seven discrete polyalanine tracts. Our previous analysis of these tracts in multiple major vertebrate clades suggested that three are mammal-specific. We now report the N-terminal HOXA13 repetitive tract structures in the monotreme Tachyglossus aculeatus (echidna). Contrary to our expectations, echidna HOXA13 possesses a unique set of polyalanine tracts and an unprecedented polyglycine tract. The data support the conclusion that the emergence of expanded polyalanine tracts in proteins occurred very early in the stem lineage that gave rise to mammals, between 162 and 315 MYA.
Keywords: HOXA13, polyalanine, monotreme, trinucleotide repeat expansion, echidna
Introduction
Homopolymeric amino acid sequences are well documented for numerous vertebrate proteins. In humans, these repetitive sequences most frequently involve glutamines, leucines, prolines, alanines or glycines (Karlin et al., 2002). The exact molecular function of homopolymeric amino acid runs are not fully understood, yet some may potentially serve as activation or repression domains (Han and Manley, 1993). Many human neurodegenerative and developmental disorders have been linked to expanded homopolymeric amino acid tracts. In particular, polyalanine tracts account for 16.9% of all human homopolymeric protein sequences (Karlin et al., 2002) and in-frame polyalanine expansions are known to cause disorders involving at least nine genes: HOXA13, HOXD13, PHOX2B, FOXL2, PABPN1, ARX, RUNX2, ZIC2, and SOX3 (Brown and Brown, 2004; Lavoie et al., 2003).
HOXA13 is a vertebrate transcription factor that is essential for proper embryonic development (McGinnis and Krumlauf, 1992; Veraksa et al., 2000). HOXA13 plays a critical role in proper embryonic patterning of the appendicular skeleton (limbs and external genitalia), the urogenital tract, as well as extraembryonic structures (placenta and umbilical artery) (Fromental-Ramain et al., 1996; Mortlock et al., 1996; Stadler et al., 2001). Over 15% of the 388aa protein is comprised of polyalanine segments, each located within the N-terminal region, preceding the homeodomain (Mortlock et al., 2000). These alanine tracts are contained in seven discrete locations in the normal human protein. Importantly, in-frame polyalanine expansions within tracts I, III, and V have all been associated with hand foot genital syndrome (HFGS), a well-characterized dominant human genetic disorder involving appendicular and urogenital structures (Goodman et al., 2000; Innis et al., 2004; Mortlock and Innis, 1997) (OMIM 140000). Understanding the evolution and the role/function of the polyalanine tracts is valuable both from the aspect of human disease, as well as novel protein function.
In our previous report, we initiated a phylogenetic study to characterize the nature of the seven polyalanine tracts throughout vertebrate evolution (Mortlock et al., 2000). Tracts I, V and VI were found to be mammal-specific, whereas the other tracts pre-dated the mammalian lineage. Monotremes are evolutionarily the most basal branching group of living mammals (protheria). Subtleties of their skull and shoulder skeletal morphology, as well as laying eggs, are general characteristics and presumably primitive for all amniotes. They lack teeth, which is homoplastic to birds, and they have mammalian synapomorphies including fur, mammary glands, and homeothermy (Grutzner and Graves, 2004). The extant members of this clade are the duck-billed platypus (Ornithorhynchus anatinus), the short-beaked echidna (Tachyglossus aculeatus), and multiple species of long-beaked echidna (Zaglossus). The polyalanine tracts in the monotreme, Tachyglossus aculeatus, were analyzed to infer the evolutionary timing of HOXA13 alanine tract expansion and correlate repeat acquisition with physiological differences in the reproductive tract.
Materials and Methods
Genomic DNA Samples and Hoxa13 Amplification
Tachyglossus aculeatus (short-beaked echidna) genomic DNA (ID# OR1448) was obtained from the Center for Reproduction of Endangered Species at the San Diego Zoo. Attempts to amplify Echidna HoxA13 N-terminal sequences in its entirety failed, thus the sequence was amplified and sequenced in overlapping segments. Tracts I–III were amplified with degenerate primers derived from opossum and mouse HoxA13 sequences: Md_A13.F 5′ GTGGCSGACGAGCTCAACAA and Md_A13.2R 5′ GGRTGCGCCATCAGGTTSCGG. Tracts IV and V were amplified with opossum derived sequences: Md_A13.2FN 5′ GCGGCCAACCAGTGCCGGAACC and 9765DR_md 5′ GACGAGCTCTGTGCGGCAGCCGAGCAGGGGCT, and tracts VI and VII were amplified last, utilizing the newly determined echidna sequence Ech_A13.3F 5′ GGGCGAACCCGTCAAGCAGTGC and our previously used mouse primer 9133D 5′ TGGTAGAAAGCAAACTCCTTGG. Echidna specific primers were made and used to amplify Ornithorhynchus anatinus (platypus) HoxA13 sequences (genomic DNA kindly provided by Evan Eichler), but no platypus-specific products were obtained.
Chelydra serpentina (snapping turtle) DNA was isolated from tail muscle of a deceased animal obtained roadside in Manchester, MI. Primers 8394D 5′ CTATGACAGCCTCCGTGCTC and Md_A13.2R were used to successfully amplify tracts I–III and Md_A13.2FN and 9765DR_md were used to successfully amplify tracts IV–V. Turtle and chicken specific primers were generated to amplify tracts VI–VII, but no turtle specific PCR products could be recovered. Spacer size results obtained for turtle tracts I–V correlate exactly with the already determined HOXA13 chicken data (Mortlock et al., 2000). This is consistent with anapsids (turtles) being more closely related to diapsids (living reptiles) than synapsids (mammalian radiation). The finding that turtle N-terminal spacers I–V are identical to those found in avian HOXA13 supports controversial data that turtles cluster with the Archosauria (Hedges and Poling, 1999), yet is in significant conflict with other trees where turtles nest with diapsids (Zardoya and Meyer, 1998). These data are included in the figures, but are not the focus of this paper.
All PCR reactions were 25 uL with a final concentration of the following: 10% glycerol, 2% formamide, 1X PWO PCR buffer (Roche Applied Science), 0.25 uM dNTPs, 0.03 U PWO polymerase (Roche Applied Science), 2 mM MgSO4, and 1 uM primers. Amplification was carried out with an initial denaturation step of 2 minutes at 97 degrees, followed by 35 cycles of 97 degrees for 30 seconds, 52 degrees for 30 seconds, and 72 degrees for 45 seconds. Final extension was carried out at 72 degrees for 5 minutes. PCR products were not directly sequenced, but instead adenine overhangs were added (dATP and Taq polymerase at 72 degrees for 30 minutes) for subsequent T/A cloning into PCR-4 TOPO (Invitrogen). These plasmid clones were then sequenced using T3 and T7 primers by the University of Michigan Sequencing Core with the addition of DMSO. To facilitate sequencing, inserts spanning tracts I–III and IV–V were further subcloned into MCS modified pBluescript (Stratagene) with either SacII or EcoRI and bidirectionally sequenced.
Hoxa13 sequences and analysis
Hoxa13 cDNA sequences (and protein translations) were obtained from Genbank: Homo sapiens NM_000522, Rattus norvegicus XM_575481, Gallus gallus NM_204139, Xenopus laevis AJ314743, Mus musculus NM_008264, Takifugu rubripes BU808376, Charina trivirgata AF083104, Typhlops richardi AF083103, Varanus dumerilli AF083102, Sceloperus undulatus AF083101, Eumeces inexpectatus AF083100, Agama agama AF083099, Monodelphis domestica AF083097, Canis familiaris AF083096, Felis catus AF083095, Oncorhynchus sp. AF107229, Gasterosteus aculeatus AF107228. Echidna and turtle nucleotide sequences generated in this report were deposited in Genbank and have accessions EF467174 and EF467175. All sequence assembly and editing was done with Editseq software from the University of Michigan site license DNAstar v6.0 sequence analysis package. Sequence alignments were done with the Megalign software using ClustalV followed by manual modification. Platypus (Ornithorhynchus anatinus) sequence assembly Oana-5.0 was utilized for evaluation of polyalanine coding sequences of numerous genes.
Results and Discussion
From Polyalanine Tracts to Heteropolymeric Spacers
Accumulation of N-terminal alanines represents the majority of the amino acid changes in HOXA13 over the course of vertebrate evolution. Hence, we have approached the evolution of the HOXA13 protein sequence primarily in terms of its seven N-terminal homopolymeric alanine tracts. While tallying only the total number of alanines in these regions does capture the majority of the phylogenetic differences of the N-terminus, there are still a number of amino acids immediately adjacent to the homopolymeric alanine tracts that are not conserved among clades (Figure 1) (Mortlock et al., 2000). The exact mechanism of HOXA13 tract expansion is unknown, but increases in the length of HOXA13 imperfect triplet repeats encoding alanine could occur via unequal crossing over or replication slippage (Innis et al., 2004; Mortlock et al., 2000). In addition, introduction/accumulation of non-alanine residues at the extremities of the tracts, which occurs in all seven domains of HOXA13 (Mortlock et al., 2000), could have happened by point mutation within pre-existing alanine codons. The question remains as to whether these additional residues have contributed to functional divergence, or whether they are merely neutral additions to the protein over millions of years of evolution.
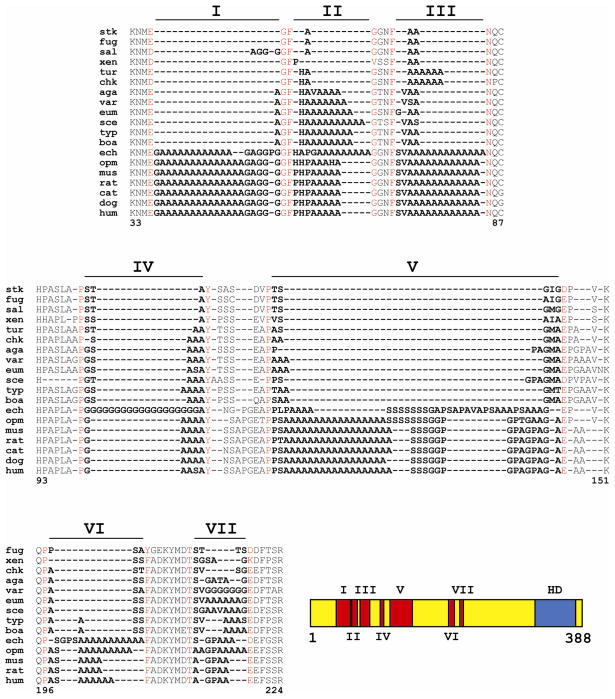
Figure 1.
(a) Phylogenetic amino acid comparison of the seven tracts in the N-terminal portion of HOXA13. Species abbreviations are as follows: stk = stickleback (Gasterosteus aculeatus), fug = fugu (Takifugu rubripes), sal = salmon (Oncorhynchus sp.), xen = xenopus (Xenopus laevis), tur = turtle (Chelydra serpentina), chk = chicken (Gallus gallus), aga = agama (Agama agama), var = varanus (Varanus dumerilli), eum = eumeces (Eumeces inexpectatus), sce = sceloperus (Sceloperus undulatus), typ = typhlops (Typhlops richardi), boa = boa constrictor (Charina trivirgata), ech = echidna (Tachyglossus aculeatus), opm = opossum (Monodelphis domestica), mus = mouse (Mus musculus), rat = rat (Rattus norvegicus), cat = cat (Felis catus), dog = dog (Canis familiaris), and hum = human (Homo sapiens). Red letters demarcate the orthologously conserved (or semi-conserved) amino acids flanking each tract. The amino acids contained within tracts (I – VII) are noted by bold lettering. The amino acid reference numbers refer to human HOXA13 protein sequence NP_000513. (b) Schematic of entire HOXA13 protein depicting placement of N-terminal tracts I through VII (in red) in relation to the C-terminal homeodomain (HD; in blue).
We have modified our traditional characterization of the homopolymeric tracts in HOXA13 to incorporate the view that these unique regions of the protein may be acting as “spacers”. This interpretation of HOXA13 N-terminal sequences stems from the idea that the alanine residues themselves may not be a direct functional component of the protein, other than simply providing necessary or optimal amino acid spacing between functional domains. In our original characterization of the human HOXA13 N-terminal region, we characterized tracts I–VII to have 14, 5, 12, 4, 18, 6, and 2 alanines respectively. In this report, we now define tract I to extend between the evolutionarily conserved KNM(E/D) motif to the invariant GF residues (Figure 1, aa 37–56), and tract II to extend between the GF residues and the next conserved G (aa 59–66). Tracts III, IV, and V extend between the invariant F and N (Figure 1, aa 71–84), the invariant P and Y (aa 100–104), and the next invariant P to the conserved E/D (aa 114–147), respectively. Tract VI is positioned between the invariant QP residues and the highly conserved FADKYMDT motif (exception in fugu: YGEKYMDT) (Figure 1, aa 198–205). The final tract, VII, is positioned between the FADKYMDT motif and the conserved E/D residue (exception in Xenopus: K) (aa 214–218). These newly established boundaries set the human complement of N-terminal amino acid spacers I–VII to 20, 8, 14, 5, 34, 8, and 5, respectively.
Evolutionary Timing and Functional Implications of Heteropolymeric Spacers
It is important to note that viewing the alanine tracts as spacers does not significantly change the evolutionary N-terminal trends of the HOXA13. In fact, the spacer residue length (as opposed to pure alanine length) appear to give a more consistent clade representation, specifically for mammals in tract II, reptiles in tract III, and both reptiles and mammals in tract IV. The trends are still replicable and tracts I, V, and VI are mammal-specific. In order to determine when these tracts arose, we characterized all of the HOXA13 spacer regions in echidna. Evaluating the size of these spacer regions in a monotreme allows for a more precise timing of the sequence expansions during the mammalian lineage. Previous analyses (Mortlock et al., 2000) suggested that the expansions occurred between the marsupial/eutherian divergence and the reptilian and bird/mammal divergence points. Inclusion of monotreme data allows us to assess an intermediate time point at 162–191 MYA (monotreme/rest of mammals) (Benton and Donoghue, 2007) (Figure 2h).
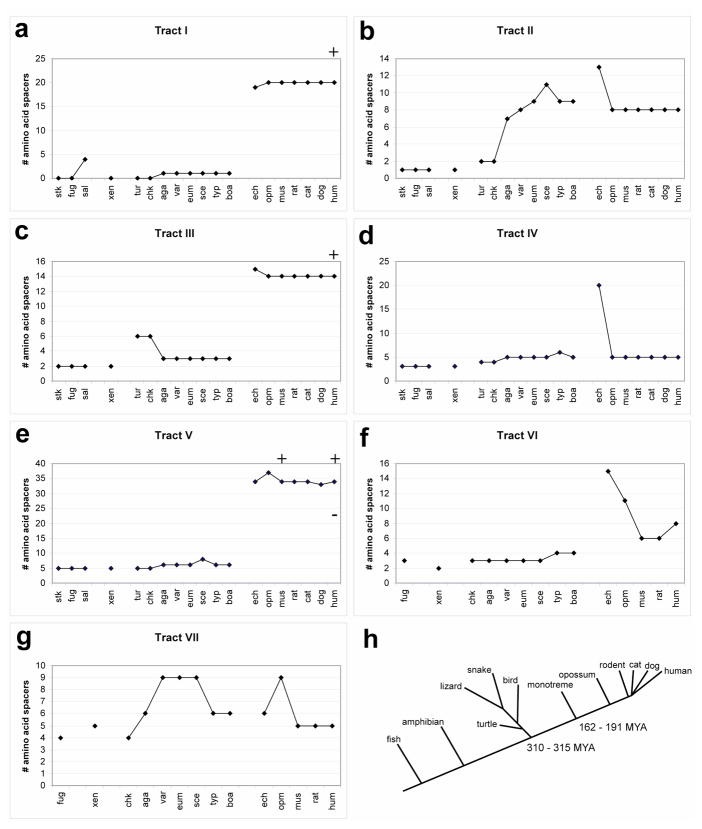
Figure 2.
(a-g) Graphs show number of amino acids contained between the phylogenetically invariant residues for each species analyzed in tracts I – VII respectively. For abbreviations see Figure 1. Data points are loosely grouped by major species clade: stickleback, fugu and salmon = fish; xenopus = amphibians; turtle, chicken, agama, varanus, eumeces, sceloporus, typhlops, boa = repiles/birds; echidna = monotremes; opossum = marsupials; mouse, rat, cat, dog, human = eutheria; (monotremes, marsupials, and eutheria are mammals). Additional human and/or mouse data points (+ or −) have been added to (a), (c), and (e) to represent the size of known human disease associated polyalanine expansions, contractions, or the engineered mouse expansion (Innis et al., 2004). (h) The schematic phylogenetic tree shows the relative divergence times (MYA = million years ago) for the major clades used in this analysis. The lengths of the lines for the various lineages are not to scale and do not necessarily reflect precise genetic distance. In our previous paper, Mortlock et al., 2000, we used an estimate (>250 MYA) of the protomammalian divergence from the reptile/bird lineage derived from Radinsky, LB, "The Evolution of Vertebrate Design" (Mortlock et al., 2000; Radinsky, 1987). It is clear from the fossil record (http://www.fossilrecord.net/dateaclade/index.html) that this divergence point is ~310–315 MYA, and our figure reflects the date from the fossil record (Benton and Donoghue, 2007). We also include the minimum/maximum constraints for the monotreme/therian split.
Based solely on these clade divergence dates (Benton and Donoghue, 2007; Radinsky, 1987), our expectation for the spacer complement in echidna was a combination of reptile/bird-like and marsupial/eutherian-like. While this a priori notion did hold true in the cases of tracts I, III, V, and VII; the echidna spacer profile overall was surprisingly unique (19-13-15-20-34-15-6) and was not merely a perfect combination of the two lineages. In the cases of tracts I, III, and V, echidna spacers were nearly identical to the other mammals (Figure 2a, c, and e). In tract I echidna has 19 residues, which is comparable to the 20 residues found in marsupials/eutherians, yet distinct from the ~1 residue spacer found in the bird/reptilian lineage. This trend is the same for tracts III and V where echidna has 15 and 34 residues, respectively, which is consistent with the other mammals (14 and ~34) and discrepant with birds and reptiles (3 and ~6). These alanine tract sizes also are consistent with the apparent maximum normal size threshold for mammalian alanine repeats, since expanded alleles cause disease (Figure 2a, c, e: +), but they also do not exceed the spacer length. In other words, while the total number of consecutive alanines is reduced, the tract length is nearly identical. Without a specific function attributed to these repeats or the neighboring conserved residues, this observation may support a model wherein a critical spacing interval is required for functional HOXA13 in mammals.
Tract VII reflects the size of either the mammals or the birds/reptiles (Figure 2g). While the size of this spacer region is not entirely consistent within or amongst clades, it is apparent that the echidna spacer size is the same as a subset of the reptiles (6) and slightly different than the eutherian mammals (5), thus making it slightly more reptile-like. It is notable that the marsupial (opossum) is the mammalian outlier for this region, as it possesses 9 spacer residues, identical to the other subset of reptiles. It is difficult to assert any functional hypothesis regarding tract VII. There is not a wide range in spacer size throughout the major vertebrate lineages (range: 4 to 9 residues), and there are no known human disease alleles associated with altered tract length in this region. Combined with the lack of strong intra-clade tract size trends, we hypothesize that tract VII does not play an important role in clade-specific HOXA13 function. However, we do not know whether size expansion was associated with a functional gain or was merely “tolerated” (Mortlock et al., 2000).
Tracts II, IV and VI possess unique spacer sizes, which for all three do not reflect HOXA13 in any species sequenced to date. Tract II has 13 residues where the rest of the mammals uniformly possess 8 and the reptiles have 7–11 (Figure 2b). Again, the functional significance of an additional 5 amino acids compared with HOXA13 in any other mammal is unknown, but in the cases of polyalanine expansions in tracts III and V an additional 6 alanines is sufficient to generate a dysfunctional protein in mice and humans (Innis et al., 2004).
In tract VI, echidna has 15 amino acid spacers (Figure 2f). This is significantly more than the number of residues in this tract in the reptiles/birds (3–4), thus making this region more similar to the mammalian profile. Interestingly, this appears to be a dynamic region of the protein within the mammalian lineage, as the number of amino acids in the spacer is variable. Echidna represents the largest spacer size (15), followed by opossum (11), humans (8) and rodents (6). In this tract early mammalian expansion was followed by contraction implying selective pressure against this in higher mammals.
Tract IV is the most novel result in that no sequenced vertebrate species has greater than 6 amino acid spacers between the evolutionarily invariant proline and tyrosine residues (Figure 1: aa 99–105), yet we show that echidna has 20 amino acids spacing these two residues (Figure 2d). In addition to the unanticipated size of this region, the content of the tract is extremely atypical of the HOXA13 N-terminal sequences we have characterized to date. Echidna tract IV consists of 19 consecutive glycine residues followed by a single alanine. This glycine run is coded for by variable length stretches of GGC codons, interspersed with GGA or GGG codons. The fact that these are imperfect triplet repeats suggests that the origin of this expanded region was most likely a multistep process likely involving a combination of point mutation, unequal meiotic recombination and/or replication slippage.
Our repeated inability to PCR amplify across the repeats from platypus genomic DNA, combined with the apparent sequencing difficulties through these regions encountered by the public draft assembly efforts (AC158254; Elliott Margulies, personal communication), suggests that the tracts in platypus are likely of the same caliber as echidna. The identification of a monotreme-specific polyglycine tract in HOXA13 lends support to the hypothesis that there may be a unique function for this domain in the monotremes.
Whether or not these collective changes in N-terminal HOXA13 are due to, or the cause of, physiological differences is yet to be determined. However, it is tempting to hypothesize that the unique echidna HOXA13 protein sequence is correlated with the distinct reproductive system of monotremes. HOXA13 is widely expressed in the developing mammalian urogenital tract in both males (epididymis, prostate, seminal vesicle, and vas deferens) and females (cervix and vagina). Moreover, HOXA13 is functional in these tissues as demonstrated by urogenital tract morphological changes with human and mouse mutant alleles (Podlasek et al., 1999; Warot et al., 1997). In addition, HOXA13 is expressed in both the umbilical artery, where it is essential for appropriate cellular stratification (Scott et al., 2005), as well as the placenta (Mortlock et al., 1996; Innis et al., manuscript in preparation). The unique, echidna-specific spacer sizes characterized for regions II, IV, and VI, identify regions of HOXA13 that may have a species-specific function in the reproductive tract of egg-laying mammals, which further evolved (change in spacer length) to contribute to reproductive tract function in placental mammals.
Importantly, the data in this report not only describe the first example of a monotreme polyalanine tract, but can also be extrapolated to define the period of widespread polyalanine tract expansion in proteins (Lavoie et al., 2003) to between 162 and 315 MYA utilizing the minimum and maximum constraints provided by analysis of the fossil record (http://www.fossilrecord.net/dateaclade/index.html) (Benton and Donoghue, 2007). To support this hypothesis, we utilized the platypus genomic sequence data to determine whether other proteins also show similar polyalanine expansions. We sought to compare the assembled platypus genomic sequences homologous to nine characterized alanine-expansion human disease genes (Brown and Brown, 2004; Lavoie et al., 2003). Of these genes (HOXA13, HOXD13, PHOX2B, FOXL2, PABPN1, ARX, RUNX2, ZIC2, and SOX3) only SOX3 was correctly assembled. In this assembled version, platypus SOX3 does not contain the polyalanine tracts present in humans. However, examination of the Ensembl gene prediction entries (GNOMON) for SOX3 retrieved XM_00111499, XM_001516070, and XM_001520731. While all three appear to be highly similar to SOX3, the latter ORF possesses two polyalanine tracts of 8 and 9 residues, yet is missing large parts of the remaining coding sequence of platypus SOX3. It is clear from this analysis that firm conclusions about the acquisition and structure of polyalanine tracts in other proteins awaits better assembly of the platypus sequence or more directed approaches such as ours. As the platypus whole-genome sequence matures a more complete analysis of polyalanine sequences for the clade will be instructive as to whether N-terminal HOXA13 is the exception or the rule for homopolymeric sequences in the monotreme lineage.
Acknowledgments
J.A.L was funded by the University of Michigan NIH Pre-Doctoral Genetics Training Program (GM 07544). We appreciate the generosity of the San Diego Zoo for provided us with Echidna genomic DNA. We value the extensive technical advice regarding DNA amplification and sequencing from both Dr. Doug Mortlock (Vanderbilt University) and Dr. Robert Lyons (University of Michigan DNA Sequencing Core).
References
- Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Brown LY, Brown SA. Alanine tracts: the expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20:51–8. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development (Cambridge, England) 1996;122:2997. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- Goodman FR, Bacchelli C, Brady AF, Brueton LA, Fryns JP, Mortlock DP, Innis JW, Holmes LB, Donnenfeld AE, Feingold M, Beemer FA, Hennekam RC, Scambler PJ. Novel HOXA13 mutations and the phenotypic spectrum of hand-foot-genital syndrome. American Journal of Human Genetics. 2000;67:197. doi: 10.1086/302961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzner F, Graves JA. A platypus' eye view of the mammalian genome. Curr Opin Genet Dev. 2004;14:642–9. doi: 10.1016/j.gde.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Poling LL. A molecular phylogeny of reptiles. Science. 1999;283:998–1001. doi: 10.1126/science.283.5404.998. [DOI] [PubMed] [Google Scholar]
- Innis JW, Mortlock D, Chen Z, Ludwig M, Williams ME, Williams TM, Doyle CD, Shao Z, Glynn M, Mikulic D, Lehmann K, Mundlos S, Utsch B. Polyalanine expansion in HOXA13: three new affected families and the molecular consequences in a mouse model. Hum Mol Genet. 2004;13:2841–51. doi: 10.1093/hmg/ddh306. [DOI] [PubMed] [Google Scholar]
- Karlin S, Brocchieri L, Bergman A, Mrazek J, Gentles AJ. Amino acid runs in eukaryotic proteomes and disease associations. Proc Natl Acad Sci U S A. 2002;99:333–8. doi: 10.1073/pnas.012608599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Debeane F, Trinh QD, Turcotte JF, Corbeil-Girard LP, Dicaire MJ, Saint-Denis A, Page M, Rouleau GA, Brais B. Polymorphism, shared functions and convergent evolution of genes with sequences coding for polyalanine domains. Hum Mol Genet. 2003;12:2967–79. doi: 10.1093/hmg/ddg329. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Innis JW. Mutation of HOXA13 in hand-foot-genital syndrome. Nature genetics. 1997;15:179. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Post LC, Innis JW. The molecular basis of hypodactyly (Hd): a deletion in Hoxa 13 leads to arrest of digital arch formation. Nature genetics. 1996;13:284. doi: 10.1038/ng0796-284. [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Sateesh P, Innis JW. Evolution of N-terminal sequences of the vertebrate HOXA13 protein. Mamm Genome. 2000;11:151–8. doi: 10.1007/s003350010029. [DOI] [PubMed] [Google Scholar]
- Podlasek CA, Clemens JQ, Bushman W. Hoxa-13 gene mutation results in abnormal seminal vesicle and prostate development. J Urol. 1999;161:1655–61. [PubMed] [Google Scholar]
- Radinsky LB. The evolution of vertebrate design. University of Chicago Press; Chicago: 1987. [Google Scholar]
- Scott V, Morgan EA, Stadler HS. Genitourinary functions of Hoxa13 and Hoxd13. J Biochem (Tokyo) 2005;137:671–6. doi: 10.1093/jb/mvi086. [DOI] [PubMed] [Google Scholar]
- Stadler HS, Higgins KM, Capecchi MR. Loss of Eph-receptor expression correlates with loss of cell adhesion and chondrogenic capacity in Hoxa13 mutant limbs. Development (Cambridge, England) 2001;128:4177. doi: 10.1242/dev.128.21.4177. [DOI] [PubMed] [Google Scholar]
- Veraksa A, Del Campo M, McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Molecular genetics and metabolism. 2000;69:85. doi: 10.1006/mgme.2000.2963. [DOI] [PubMed] [Google Scholar]
- Warot X, Fromental-Ramain C, Fraulob V, Chambon P, Dolle P. Gene dosage-dependent effects of the Hoxa-13 and Hoxd-13 mutations on morphogenesis of the terminal parts of the digestive and urogenital tracts. Development (Cambridge, England) 1997;124:4781. doi: 10.1242/dev.124.23.4781. [DOI] [PubMed] [Google Scholar]
- Zardoya R, Meyer A. Complete mitochondrial genome suggests diapsid affinities of turtles. Proc Natl Acad Sci U S A. 1998;95:14226–31. doi: 10.1073/pnas.95.24.14226. [DOI] [PMC free article] [PubMed] [Google Scholar]