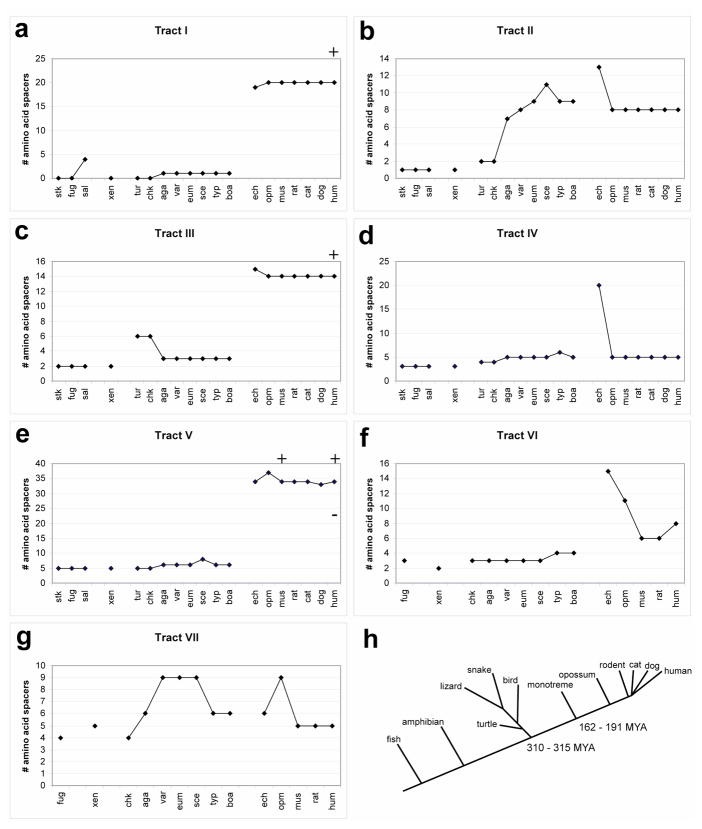
Figure 2.
(a-g) Graphs show number of amino acids contained between the phylogenetically invariant residues for each species analyzed in tracts I – VII respectively. For abbreviations see Figure 1. Data points are loosely grouped by major species clade: stickleback, fugu and salmon = fish; xenopus = amphibians; turtle, chicken, agama, varanus, eumeces, sceloporus, typhlops, boa = repiles/birds; echidna = monotremes; opossum = marsupials; mouse, rat, cat, dog, human = eutheria; (monotremes, marsupials, and eutheria are mammals). Additional human and/or mouse data points (+ or −) have been added to (a), (c), and (e) to represent the size of known human disease associated polyalanine expansions, contractions, or the engineered mouse expansion (Innis et al., 2004). (h) The schematic phylogenetic tree shows the relative divergence times (MYA = million years ago) for the major clades used in this analysis. The lengths of the lines for the various lineages are not to scale and do not necessarily reflect precise genetic distance. In our previous paper, Mortlock et al., 2000, we used an estimate (>250 MYA) of the protomammalian divergence from the reptile/bird lineage derived from Radinsky, LB, "The Evolution of Vertebrate Design" (Mortlock et al., 2000; Radinsky, 1987). It is clear from the fossil record (http://www.fossilrecord.net/dateaclade/index.html) that this divergence point is ~310–315 MYA, and our figure reflects the date from the fossil record (Benton and Donoghue, 2007). We also include the minimum/maximum constraints for the monotreme/therian split.