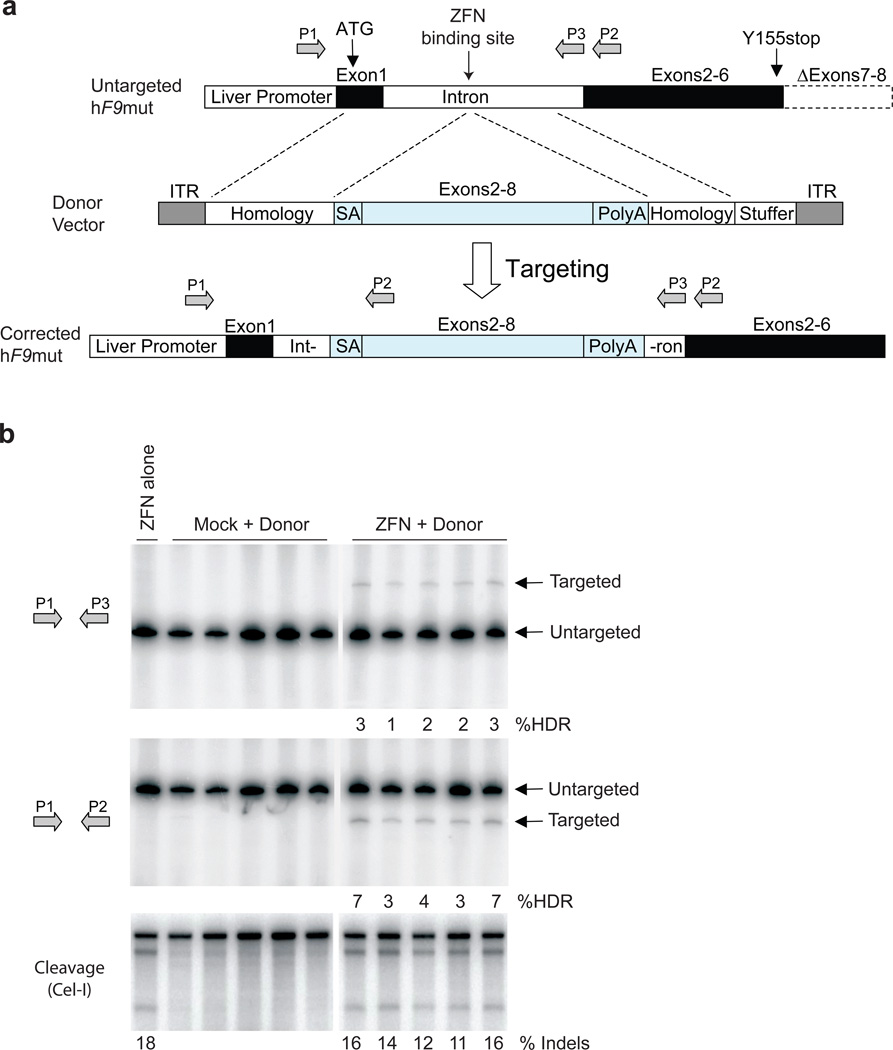
Figure 3. F9 ZFNs promote AAV-mediated targeting of wild-type F9 exons 2–8 to hF9mut intron 1 in vivo.
a, The hF9mut gene mutation (truncation of exons 7&8) can be bypassed by targeted integration of hF9 exons 2–8 into intron 1. Targeted and untargeted hF9mut alleles can be differentiated through PCR using primers P1, P2, and P3. Location of methionine start codon, and premature stop mutation indicated by arrows. The left arm of homology spans from the beginning of exon 1 to the ZFN target site. (Deletion of exon 1 from left homology arm does not alter results, Supplementary Figure 13). The right arm of homology spans intronic sequence 3’ of the ZFN target site. b, PCR analysis with primer pairs P1/P3 (upper panel) and P1/P2 (middle panel) demonstrating successful gene targeting by HDR upon I.P. co-injection of 5e10 v.g. AAV8-ZFN and 2.5e11 v.g. AAV8-Donor in hF9mut/HB mice at day 2 of life (n=5), but not with injection of 5e10 v.g. AAV8-ZFN alone (n=1), or co-injection of 5e10 v.g. AAV8-Mock and 2.5e11 v.g. AAV8-Donor (n=5). Mock vector replaces F9 ZFN coding sequences with Renilla Luciferase. PCR was performed using 32P-labeled nucleotides, followed by PAGE and product band intensity quantification by autoradiography to evaluate targeting frequency. Targeting frequencies are rounded down to the nearest whole number. Lower panel. IP injection of AAV8-ZFN expression vector into hF9mut mice results in cleavage of intron 1. The Cel-I assay was performed on liver DNA to determine the frequency of ZFN-induced insertions and deletions (indels), indicated as the % Indels at the base of each lane, resulting from cleavage of the hF9mut intron. Lanes with no quantification had no detectable HDR or indels. Each lane represents an individual mouse.