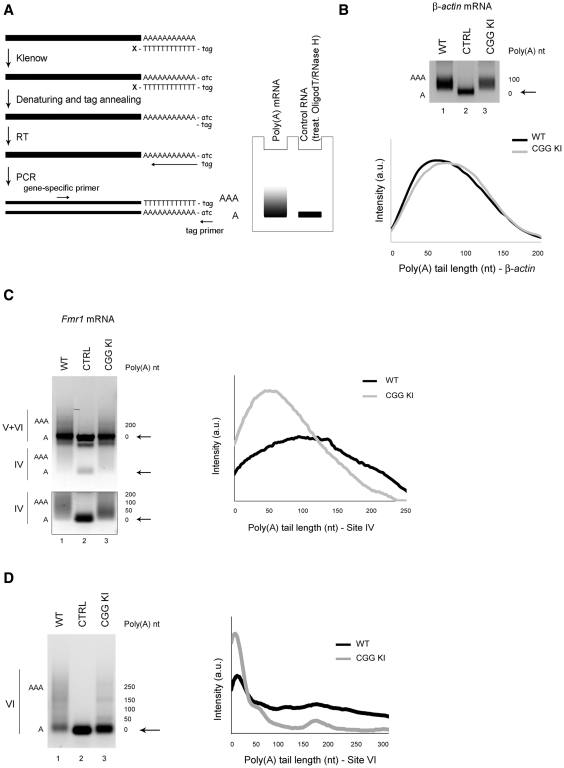
Figure 4.
Polyadenylation state of Fmr1 mRNA variants in the CGG KI mouse brain. (A) Left panel: schematic view of the polyadenylation assay (PAT). According to di Penta et al. (42), the poly(A) tails are tagged by incubating the RNA with a (T)12-tag oligonucleotide, blocked at the 3′-end, in the presence of dNTPs and Klenow enzyme to fill in the complementary tag sequence. The RNA is then denatured and annealed to a DNA primer, identical to the tag, to start a reverse transcription (RT). The cDNA is then amplified using a gene-specific forward primer and the reverse tag oligo. Right panel: cartoon of a polyadenylation profile obtained with a PAT assay. The PCR of a polyadenylated mRNA gives rise to a smear while the same mRNA deadenylated with oligodT and RNase H prior to poly(A) tagging is used as a negative control and gives a sharp band. (B) Upper panel: β-actin mRNA in WT (lane 1) and CGG KI (lane 3). Deadenylated RNA is shown as negative control (lane 2) and the deadenylated form is indicated by black arrows. Lower panel: dispersion graph representing the distribution of the β-actin polyadenylated transcripts in WT (black line) and CGG KI (grey line). The signal intensity along the lane has been plotted against the poly(A) tail length, estimated from the molecular markers loaded on the same gel. (C) PAT for all three poly(A) Fmr1 mRNA variants. Because of close proximity, the transcripts containing sites V and VI cannot be discriminated and therefore they are not taken into exam (upper panel). Black arrows points to the deadenylated form. The polyadenylation of transcripts using site IV from WT (lane 1) and CGG KI (lane 3) has been independently acquired and highlighted in the box below. Deadenylated RNA treated as mentioned above, is shown as negative control (lane 2). Right panel: dispersion graph for Fmr1 variants using site IV in WT (black line) and CGG KI (grey line). (D) Left panel: PAT for Fmr1 variants using site VI in WT (lane 1) and CGG KI (lane 3) brain. Deadenylated RNA as above is used as negative control (lane 2). Right panel: Dispersion graph representing the distribution of the polyadenylated transcripts using site VI in WT (black line) and CGG KI (grey line).