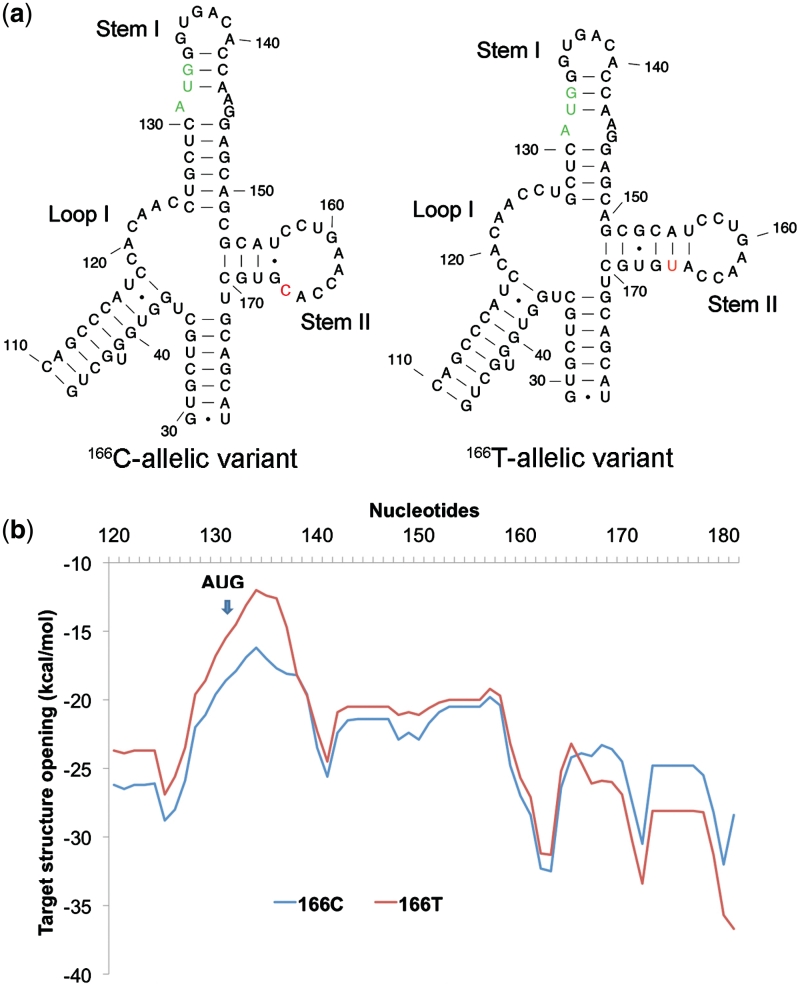
Figure 2.
Secondary structure analysis of 166C and 166T-allelic variants of COMT by Mfold. (a) Free energy profile (ΔG) for target structure opening in the vicinity of the rs4633 SNP and the start codon for 166C and 166T-allelic variants determined using OligoWalk. (b) The most probable RNA secondary structural predictions for 166C and 166T-allelic variants near start codon. We zoom in toward the regional differences between the allelic variants. The start codon is colored in green and the varying SNP is colored in red. The regions of difference are within the motifs labeled Loop I, Stem I and Stem II. The two stems leading to the Loop I and Stem II (capped at base pairs 46–110 and 29–178, respectively) are identical as are the rest of the nucleotides within the 210-nt window (Supplementary Data, Figures S1–S3). Both DMD and Mfold predict identical secondary structures for the 166C-allelic variant and 166T-allelic variant.