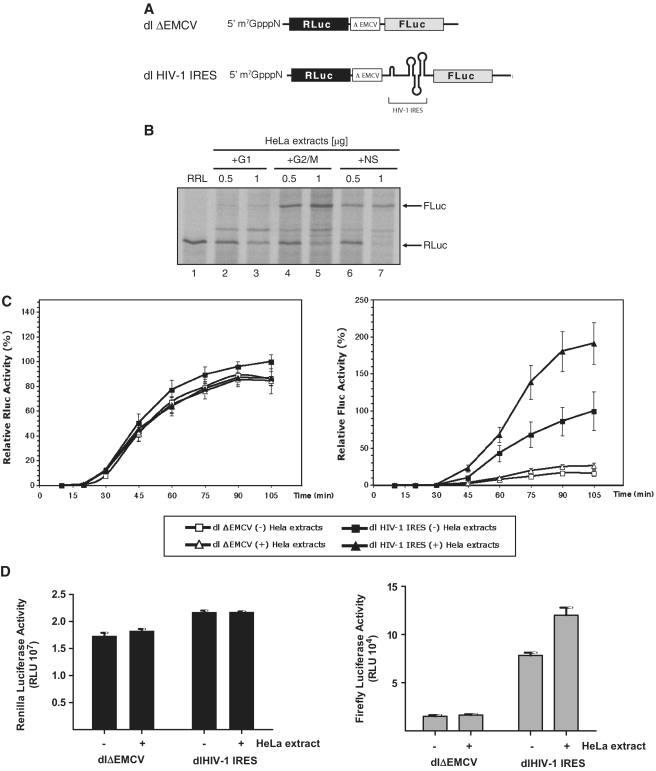
Figure 1.
HeLa cell cytoplasmic factors are required for HIV-1 IRES activity. (A) Schematic representation of the dl HIV-1 IRES and dlΔEMCV RNAs used in this study. (B) RRL alone or supplemented with cytoplasmic extracts (0.5 or 1 µg of total protein) generated from NS HeLa cells or cells arrested in G1 or in G2/M were programmed with the dl HIV-1 IRES RNA. [35S]-methionine-labeled proteins were resolved by SDS–PAGE and visualized as indicated in ‘Materials and Methods’ section. (C) Kinetics of HIV-1 IRES translation in the presence of G2/M HeLa extracts. The capped dlΔEMCV (open shapes) or dl HIV-1 IRES (filled shapes) RNAs (8 ng/µl) were used to program RRL. In vitro translation reactions were supplemented with 160 ng/µl of G2/M cytoplasmic extracts (Δ). Renilla luciferase (RLuc) and Firefly luciferase (FLuc) activities were measured at the indicated times. The RLuc and FLuc activities of the dl HIV-1 IRES measured at 105 min of in vitro translation in non-suplemented RRL (filled square) were arbitrary set to 100%. Relative RLuc activity (left panel) and relative FLuc activity (right panel) are shown. Values are the means ± SEM (error bars) of five independent experiments. (D) Capped and polyadenylated RNA corresponding to the dlΔEMCV or dl HIV-1 IRES vectors (6.25 ng) were microinjected into X. laevis oocytes with (+) or without (−) cytoplasmic extracts generated from G2/M arrested HeLa cells (200 ng) as described in ‘Materials and Methods’ section. Oocytes were harvested 24 h after the microinjection and processed and RLuc and FLuc activities were determined RLU. The RLuc (left panel) and FLuc (right panel) activities for each RNA are shown. Each value is the mean ± SEM from at least three oocytes obtained from different animals.