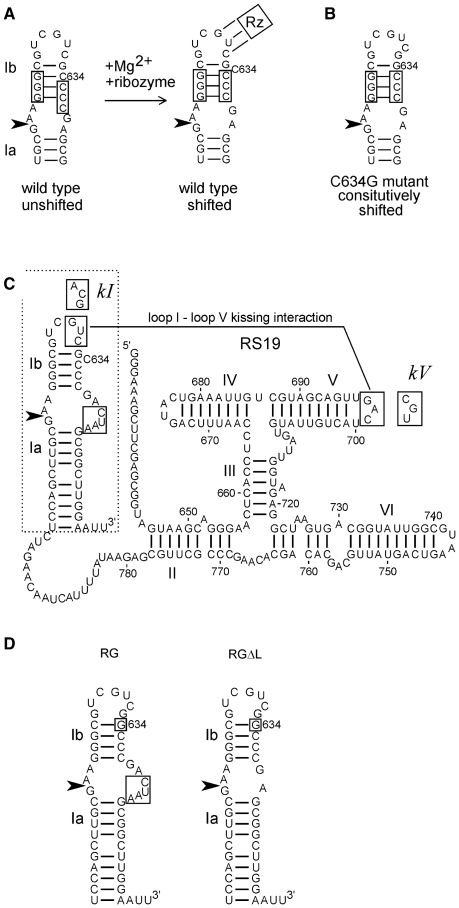
Figure 1.
Sequences and structures of VS ribozyme constructs. (A) The secondary structure of helix Ib and the internal loop containing the cleavage–ligation site (arrowhead) rearranges during Mg2+-dependent folding with the rest of the ribozyme (represented by Rz; the three lines connecting loop I to Rz represent the loop I–loop V kissing interaction [see Figure 1C and ref. (18,20)]. (B) The C634G mutant constitutively adopts the shifted conformation required for activity. (C) Secondary structure diagram of the RS19 version of the VS ribozyme, drawn with helix I in the shifted conformation (with C634 bulged out of the helix). Nucleotide numbers correspond to full-length VS RNA; helices are numbered with Roman numerals; the cleavage–ligation site is indicated by the arrowhead (19,38). The non-natural four extra nucleotides in the 3′ side of the cleavage–ligation site loop are boxed [see ref. (19) for details]. Boxed sequences in loops I and V interact via a kissing interaction indicated by the solid line (9). Mutant loop I and V sequences used to construct the kI, kV and kI+V mutants are indicated in boxes adjacent to their natural loop sequences. (D) The region of the RS19 ribozyme (dashed outline in C) into which the C634G mutation (boxed) was introduced to construct the RG ribozyme; deletion of the four non-natural nucleotides in the cleavage site loop of RG produced the RGΔL ribozyme.