Abstract
NLR family proteins play important roles in innate immune response. NOD1 (NLRC1) activates various signaling pathways including NF-κB in response to bacterial ligands. Hereditary polymorphisms in the NOD1 gene are associated with asthma, inflammatory bowel disease, and other disorders. Using a high throughput screening (HTS) assay measuring NOD1-induced NF-κB reporter gene activity, followed by multiple downstream counter-screens that eliminated compounds impacting other NF-κB pathways, 2-aminobenzimidazole compounds were identified that selectively inhibit NOD1. Mechanistic studies of a prototypical compound, Nodinitib-1 (ML130; CID-1088438), suggest these small molecules cause conformational changes of NOD1 in vitro and alter NOD1 subcellular targeting in cells. Altogether, this inaugural class of inhibitors provides chemical probes for interrogating mechanisms regulating NOD1 activity and tools for exploring the roles of NOD1 in various infectious and inflammatory diseases.
Keywords: NOD1, NOD2, NLR, NF-κB, HTS
INTRODUCTION
NLRs (NACHT and Leucine Rich Repeat domain containing proteins) constitute a prominent family of innate immunity proteins found in mammals (Kanneganti et al., 2007; Ting et al., 2008). NLR family members, NOD1 (NLRC1, CARD4 or CLR7.1) and NOD2 (NLRC2, also CARD15; CD; BLAU; IBD1; PSORAS1; CLR16.3) are of particular interest, because they recognize distinct structures derived from bacterial peptidoglycans and activate various signaling pathways important for host defense and inflammation, including NF-κB, stress kinases, and Interferon response factors (IRFs) (Pandey et al., 2009; Strober et al., 2006; Watanabe et al., 2010).
NOD1 gene polymorphisms are associated with several human inflammatory disorders, including sarcoidosis, Crohn’s disease, asthma, and autoimmune uveitis (Carneiro et al., 2008; Eckmann and Karin, 2005; Franchi et al., 2008; Strober et al., 2006; Tattoli et al., 2007). Recently, NOD1 has additionally been implicated in neuroinflammatory processes involved in the progression of multiple sclerosis (MS) (Shaw et al., 2011). In an animal model of MS (experimental autoimmune encephalomyelitis), Nod1−/− mice show markedly reduced incidence of hind-limb paralysis, decreased axon demyelination, and diminished neuroinflammatory infiltrates (Shaw et al., 2011). NOD1 has also been implicated in vascular inflammation. NOD1 activation stimulates cytokine production by human coronary artery endothelial cells in culture. Moreover, administration of NOD1 agonists into mice induces coronary arteritis, with dense cellular infiltrates (i.e. neutrophils and macrophages), showing histopathological similarity to the acute phase of Kawasaki disease - a rare childhood disease characterized by inflammation of blood vessels (vasculitis) (Nishio et al., 2011). A mouse model of NOD1-induced ocular inflammation (uveitis) has also been reported (Rosenzweig et al., 2009). Thus, access to chemical NOD1 inhibitors should empower research on defining the role of this NLR family protein in numerous acute and chronic inflammatory diseases, allowing for an exploration of whether novel therapeutic interventions based on targeting this class of proteins are feasible.
Here we describe the identification of a series of 2-aminobenzimidazole derivatives that selectively inhibit NOD1. We have developed and optimized a cell-based high throughput screening (HTS) assay platform in which either NOD1 or NOD2 activation stimulates a NF-κB-responsive luciferase reporter gene. Various downstream counter-screens, combined with insights provided by cheminformatics analysis, were further applied to validate the 2-aminobenzimidazole scaffold as an inaugural family of NOD1-specific inhibitors.
RESULTS & DISCUSSION
HTS campaign identifies NOD1 inhibitors
We devised cell-based HTS assays utilizing a NF-κB-driven luciferase reporter gene as a measure of NOD1 or NOD2 activity. For the NOD1 assay, HEK293T cells were stimulated with NOD1 ligand, Ala-γ-Glu-diaminopimelic acid (γ-tri-DAP) (Chamaillard et al., 2003; Girardin et al., 2003), a component of peptidoglycan (PG), relying on endogenous NOD1 expression to result in NF-κB reporter gene activation (PubChem AID 1578). The Z’ values for the optimized assay performed in either 384 or 1536 well format were consistently in the range of 0.67 to 0.73. The NOD2 assay utilized stable over-expression of NOD2 in HEK293T cells, which employed the same NF-κB luciferase reporter gene and which was also optimized to Z’ factor > 0.5 in both 384- and 1536-well formats (PubChem AID 1566).
The NIH library (~300,000 compounds) was screened at an average concentration of ~4 μM using the NOD1 and NOD2 HTS assay in 1536 well format to identify candidate inhibitors based on NF-κB reporter gene activity (Suppl. Figs. 1 and 2). Hits were counter-screened to eliminate cytotoxic compounds (false-positives), and using cheminformatic filters to eliminate historically promiscuous bioactives, 2,481 hits were identified that inhibited either NOD1, NOD2 or both (Suppl. Fig. 3A). These hits were then further tested at the same concentration against the same HEK293T-NF-κB luciferase cells stimulated with TNFα to induce NF-κB by an alternative means (PubChem AID 1852), thus eliminating 1,286 non-specific compounds. The hit compounds were re-ordered and re-tested, reducing the confirmed hits to 536 compounds. Testing of these compounds in dose-response experiments using both NOD1 and NOD2 NF-κB reporter gene assays revealed 309 with IC50 ≤ 10 μM and with little or no cytotoxicity at 20 μM (PubChem AID 2335). Counter-screening the NOD1 and NOD2 hits against each other revealed 183 compounds showing ≥10-fold target selectivity of NOD1 over NOD2 (Suppl. Fig. 3B). Compounds that inhibited NOD2 will be described elsewhere.
Pathway selectivity assays reveal NOD1-selective inhibitors
Several cell-based assays were developed to differentiate compounds that inhibit NF-κB induction by other upstream activators from NOD1/NOD2 selective compounds. For instance, using the same HEK293T-NF-κB-luciferase cells, we compared the ability of compounds to suppress NF-κB activity induced by NOD1 ligand (γ-tri-DAP), NOD2 ligand (muramyl dipeptide [MDP]), TNFα, protein kinase C activators (phorbol myristic acetate [PMA] and ionomycin), and DNA damaging agents (doxorubicin) (Fig. 1A-B). Consistently, a series of 2-aminobenzimidazole derivatives inhibited NF-κB activation only after γ-tri-DAP treatment (Figs. 1A-B), thus showing promise as potential NOD1-specific NF-κB inhibitors. A structure activity relation (SAR) analysis was initiated by both analog-by-catalog approach and internal medicinal chemistry effort, wherein a total of 78 compounds were synthesized. Interestingly, we observed that the presence of 2-amino and sulfonamide functionality on the benzimidazole ring is pertinent for the bioactivity of these compounds (Suppl. Table 1). In general, electron-donating groups on the aromatic ring resulted in more potent and selective compounds. A detailed description of SAR and medicinal chemistry effort will be described separately.
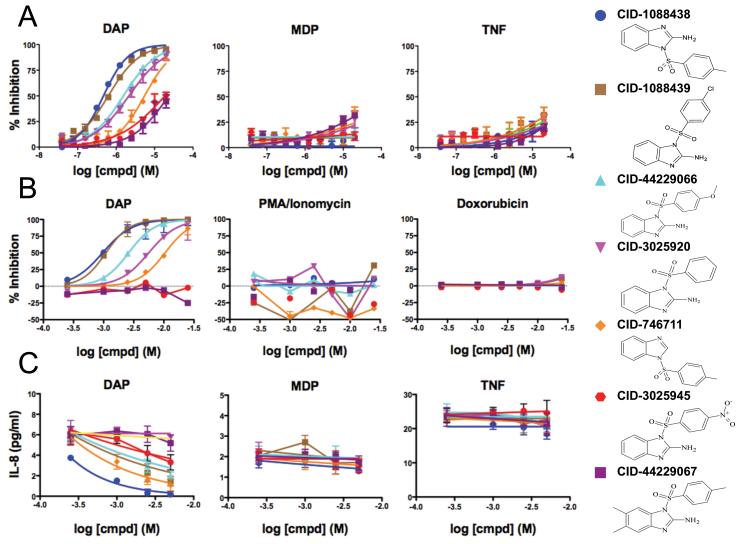
Figure 1. Profiling 2-aminobenzimidazole derivatives for pathway selectivity.
(A) HEK 293T NF-κB-luciferase cells (6,000 cells/well in 1536-well plate) were treated with 0.75 μg/ml γ-tri-DAP, 0.7 μg/ml MDP, or 0.1 ng/mL TNFα for 14 hours, in the presence of increasing amounts of the selected compounds. Luciferase activity data were used to calculate percent inhibition (mean ± standard error [SEM], n=2). (B) NF-κB-luciferase-containing cells were treated γ-tri-DAP (5 μg/ml), 0.4 μg/ml Doxorubicin, or 10 ng/ml PMA/Ionomycin for 18 hours (10,000 cells/well in 384-well plate), measuring luciferase activity and calculating % inhibition (mean ± standard deviation [SD], n=3). (C) MCF-7 cells stably expressing NOD1, NOD2, or GFP (50,000 cells/well in 96-well plate) were treated with 5 μg/ml γ-tri-DAP, 5 μg/ml MDP or 5 ng/ml TNFα, supplemented with 1.5 μg/ml cycloheximide, for 24 hours, in the presence of increasing amounts of the selected compounds (in micromolar, μM). IL-8 levels were quantified by ELISA. Data represent mean ± SD, n=3. 2-D structures of selected compounds are shown (see Suppl. table 1 for complete SAR). See also Figures S1-S5, and S7.
To extend the analysis of the candidate NOD1 inhibitors beyond reporter gene assays, we also measured the levels of a NF-κB-inducible cytokine, interleukin-8 (IL-8). Using a recently described assay employing breast cancer MCF-7 cells over-expressing NOD1 or NOD2 (da Silva Correia et al., 2006), we measured IL-8 secretion into culture supernatants following stimulation with NOD1 ligand (γ-tri-DAP), NOD2 ligand (MDP), or TNFα (Fig. 1C). Again, 2-aminobenzimidazole series compounds selectively inhibited IL-8 production induced by NOD1 ligand but not other stimuli, with compound CID-1088438 representing the most potent of the analogs. Cell viability was not affected by CID-1088438 under diverse cell treatments (Suppl. Fig. 4). CID-1088438 also inhibited γ-tri-DAP-induced expression of the prototypical NF-κB target gene IκBα at the mRNA level (Suppl. Fig. 5). Indeed, CID-1088438 inhibits γ-tri-DAP-dependent activation of NF-κB (IκBα phosphorylation and degradation) and MAPK (p38 phosphorylation) signalings, without affecting Akt survival pathway (Suppl. Fig. 6), consistent with the known roles of NOD1 in signal transduction (Allison et al., 2009; Inohara et al., 1999; Opitz et al., 2006; Tattoli et al., 2007).
In addition to NLRs, Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) constitute important families of pathogen receptors (Creagh and O’Neill, 2006). Human myelomonocytic THP-1 cells containing a NF-κB/AP-1-inducible reporter gene encoding secreted alkaline phosphatase (SEAP) were employed for convenient monitoring of NF-κB activity. After inducing differentiation with PMA, THP.1 macrophages were treated for 24 hours with CID-1088438 or its inactive analog (CID-44229067, Suppl. Table 1) and various TLR agonists, assessing effects on NF-κB-inducible SEAP activity. No inhibitory effects of CID-1088438 were observed for any of the TLR agonists tested (TLR1, 2, 4, 5, 6 and 8) (Fig. 2A). Note that while the NOD1 ligand γ-tri-DAP is a weak inducer of NF-κB activity in THP.1 macrophages, inhibition by CID-1088438 was highly reproducible.
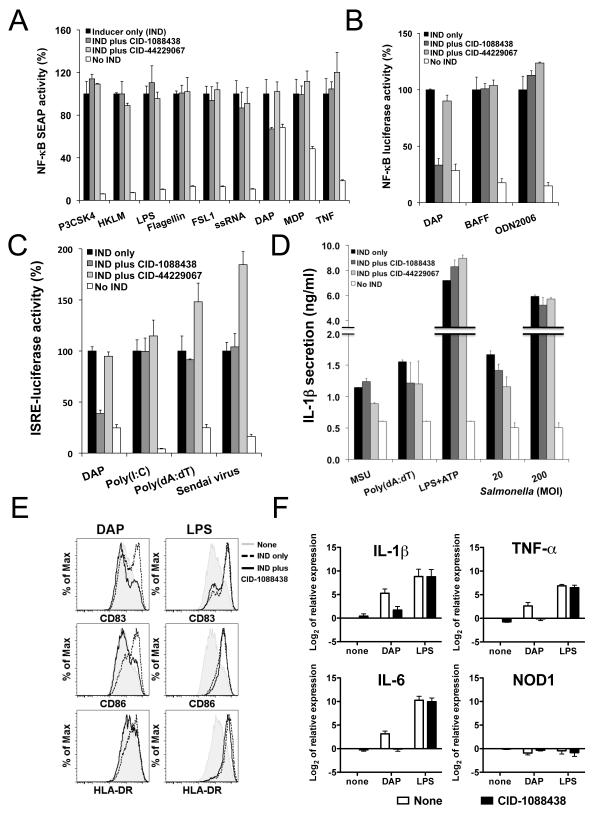
Figure 2. CID-1088438 specifically inhibits NOD1-dependent signaling pathways.
(A) PMA-differentiated THP.1-cells containing NF-κB-driven SEAP (105 cells/well in 96-well plate) were cultured with or without 5 μM CID-1088438 or CID-44229067 and various TLR/NLR inducers: 0.5 μg/ml Pam3CSK4 (TLR1/TLR2), 5×107 cells/ml HKLM (TLR2), 1 μg/ml FSL-1 (TLR6/2), 0.5 μg/ml LPS (TLR4), 0.5 μg/ml Flagellin (TLR5), 1 μg/ml ssRNA40 (TLR8), 5 μg/ml γ-tri-DAP (NOD1), 5 μg/ml MDP (NOD2) or 5 ng/ml TNFα. After 24h incubation, SEAP activity in culture supernatants was measured, expressing data as percentage relative to treatment with inducer only (indicated as 100%; mean ± SEM, n=2). (B) 697 cells stably containing a NF-κB-luciferase reporter gene (105 cells/well in 96-well plate) were cultured with or without 10 μM CID-1088438 or CID-44229067, in combination with 20 μg/ml γ-tri-DAP, 100 ng/ml BAFF or 5 μM ODN2006 (TLR9). Luciferase activity was measured 24h later (mean ± SD; n=3). (C) 293T cells, stably expressing luciferase reporter gene driven by IFN responsive elements (105 cells/well in 96-well plate), were cultured with or without 5 μM CID-1088438 or CID-44229067, in combination with 10 μg/ml γ-tri-DAP (NOD1), 1 μg/ml poly(I:C) with lipid transfection (LyoVec) (RIG-I/MDA-5), 1 μg/ml Poly(dA:dT) (LyoVec) (IRF3) or Sendai Virus (classical IRF3 inducer). Luciferase activity was measured after 24 hrs (mean ± SD; n=4). (D) RAW264.7 cells (5×104 cells/well in 96-well plate) were treated with 5 μM of CID-1088438 or CID-44229067, then stimulated with 100 ng/ml monosodium urate (MSU), 1 μg/ml poly(dA:dT) or 1 μg/ml LPS plus 5 mM ATP, after LPS pre-treatment (induction of pro-IL-1β synthesis), or infected with S. typhimurium at multiplicity of infection (MOI) of 20 and 200 bacteria per mammalian cell. Supernatants were collected after either 2 hrs (Salmonella infection) or 4 hrs (all others) and IL-1β levels were quantified by ELISA (mean ± SD; n=3). (E) Dendritic cells (DCs) were activated with either 5 μg/ml γ-tri-DAP or 100 ng/ml LPS, in the presence or absence of 15 μM CID-1088438. After 24hr, flow cytometry analysis was performed for CD83, CD86 and HLA-DR markers. Representative data from one donor are shown (n=3). (F) Expression of prototypical NF-κB target genes in primary monocyte-derived DCs. Cells were treated with either 5 μg/ml γ-tri-DAP or 100 ng/ml LPS, in the presence or absence of 15 μM CID-1088438. After 4hr, relative mRNA expression of IL-1β, IL-6, TNFα and NOD1 were determined by quantitative PCR. Results were normalized according to β-actin levels (mean ± SEM of three donors). See also Figures S1-S11.
A non-canonical NF-κB pathway stimulated by certain TNF family members, including B cell-activating factor (BAFF), involves ubiquitin-mediated processing of the NF-κB subunit p100 to p52 and is dependent on phosphorylation of p100 by IKK subunit, IKK1 (or IKKα) (Claudio et al., 2002; Tergaonkar, 2006). Using 697 pre-B leukemia cells containing a NF-κB-luciferase reporter gene, we verified that the non-canonical NF-κB activation induced by BAFF (Claudio et al., 2002) is not inhibited by CID-1088438 (Fig. 2B), whereas NF-κB activity induced by NOD1 ligand γ-tri-DAP is inhibited. CID-1088438 also did not inhibit NF-κB activity induced by TLR9 ligand CpG DNA in these cells (Fig. 2B).
The RIG-I like receptors (RLRs) comprise a family of cytoplasmic RNA helicases that include RIG-I (retinoic-acid-inducible protein I), and MDA-5 (melanoma-differentiation-associated gene 5), implicated in viral double-strand RNA recognition (Creagh and O’Neill, 2006). RIG-I and MDA-5 bind the mitochondrial membrane protein MAVS to initiate a signaling cascade that includes induction of the type I interferon response (Seth et al., 2006). In addition to stimulating NF-κB, NOD1 also binds MAVS to stimulate interferon (IFN) production by activating IRFs (Watanabe et al., 2010). Using HEK293T cells stably containing an IFN-sensitive response element (ISRE)-driven luciferase reporter gene, we tested the effects of compound CID-1088438 on several IFN inducers, including NOD1 ligand γ-tri-DAP, poly(I:C), poly(dA:dT), and a RNA virus (Sendai virus). While CID-1088438 suppressed ISRE- driven reporter gene activity induced by γ-tri-DAP, no inhibition was observed for the other interferon response stimuli (Fig. 2C). In contrast, the inactive analog CID-44229067 did not inhibit γ-tri-DAP-induced ISRE reporter gene activity (Fig. 2C). These results further confirm the selectivity of the NOD1 inhibitory 2-aminobenzimidazole compounds, and also indicate that they act upstream of the divergence of the NF-κB and IFN-dependent pathways activated by NOD1.
Many NLRs form complexes with caspase-1, creating so-called “inflammasomes” responsible for proteolytic processing of inflammatory cytokine interleukin 1-beta (IL-1β) (Martinon and Tschopp, 2007; Petrilli et al., 2007). CID-1088438 did not inhibit IL-1β secretion induced by various inflammasome activators. (Fig. 2D and suppl. Fig. 7), indicating a lack of promiscuity towards other NLRs.
CID-1088438 selectively inhibits responses of primary dendritic cells to NOD1 ligand
To extend the analysis of CID-1088438 beyond immortalized cell lines to primary cells, we performed experiments using ex vivo cultures of human monocyte-derived dendritic cells (DC). DCs were activated with either γ-tri-DAP or lipopolysaccharide (LPS), in the presence or absence of active compound CID-1088438. CID-1088438 reduced cell surface expression of co-stimulatory molecules CD83, CD86 and HLA-DR (Fig. 2E) and also inhibited expression of IL-1β, IL-6 and TNFα (Fig. 2F) elicited by γ-tri-DAP (but not by LPS), without causing cytoxicity. No significant changes in NOD1 expression levels were observed (Fig. 2F).
Mechanisms of chemical inhibitors of NOD1
NOD1 activates NF-κB in partnership with various interacting proteins, particularly RIP2, IAPs, and IKKγ/NEMO (Bertin et al., 1999; Hasegawa et al., 2008; Inohara et al., 1999; Krieg et al., 2009; Stehlik et al., 2003), wherein NOD1 binds directly to RIP2, which in turn interacts with IAPs, forming a complex that stimulates IKK activation (Krieg and Reed, 2010). Gene transfection experiments suggested that CID-1088438 targets NOD1 signaling upstream of RIP2 (Fig. 3A). No significant impact of the compound was observed in cells over-expressing IKKγ/NEMO, MYD88, FLIP, CARD6, APAF1 or NLRC4 (data not shown), confirming specificity.
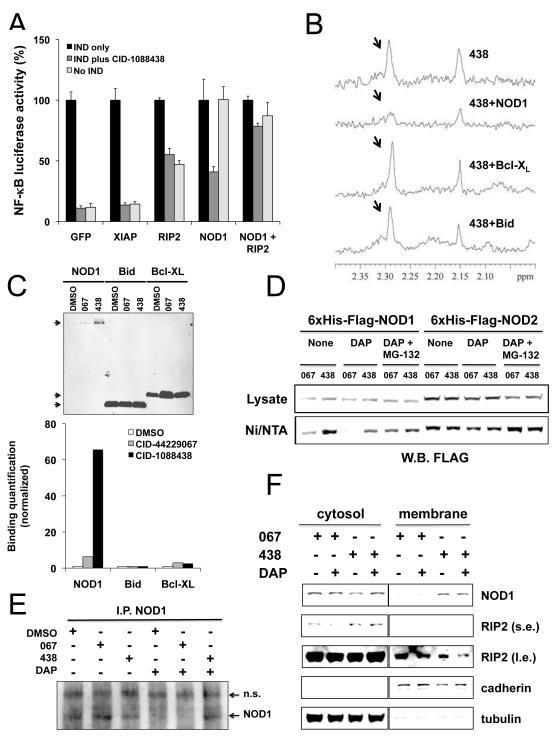
Figure 3. Analysis of mechanisms of chemical inhibitors of NOD1.
(A) HEK 293T-NF-κB-luciferase cells were transfected with plasmids encoding NOD1, RIP2, XIAP, or GFP in various combinations. After 24hr, cells were cultured with or without 3.5 μg/ml γ-tri-DAP, 10 μM of CID-1088438, or combinations of these reagents for 24 hrs before measuring luciferase activity normalizing data to GFP-transfected cells (100%) (mean ± SD, n=4). (B) 1D 1H-NMR spectra were collected for CID-1088438 (50 μM) in the absence (upper spectrum) and presence (lower spectra) of 5 μM of His6-Flag-NOD1, His6-Bcl-XL, or His6-Bid purified proteins, respectively. CID-1088438-derived proton signal intensity (arrows) is only suppressed in the presence of NOD1, thereby demonstrating direct interaction between ligand and protein. (C) Purified His6-Flag-NOD1, His6-Bcl-XL, or His6-Bid proteins (≈0.4 ug) were pre-incubated for 60 minutes with 20 μM CID-1088438 or CID-44229067 (DMSO as control). Ni-NTA agarose beads were then added and the mixture was incubated overnight at 4°C. Ni/NTA pull-down was performed and samples were analyzed by immunoblotting using anti-His antibody (top). Quantification of proteins on blots was performed, normalizing data relative to DMSO control (bottom). (D) MCF-7 cells stably expressing His6-FLAG-NOD1 or His6-FLAG-NOD2 were cultured with 5 μM of CID-1088438 or CID-44229067 and 10 μg/ml γ-tri-DAP alone or with 1 μM MG-132. After 16hr, cells were lysed and equal amounts of protein samples were pulled down using Ni/NTA resin beads. Total protein lysates (30 μg) and Ni/NTA-bound proteins were analyzed by immunoblotting using anti-FLAG antibody. (E) HCT-116 cells were treated with DMSO (control) or with 10 μg/ml γ-tri-DAP, with or without 5 μM of CID-1088438 or CID-44229067. After 24 hr in rabbit anti-NOD1 antibody immunoprecipitates were prepared and analyzed by immunoblotting using rat anti-NOD1 antibody. Non-specific (n.s.) and NOD1-specific bands are indicated. (F) MCF-7 cells stably expressing His6-FLAG-NOD1 were cultured with 5 μM of CID-1088438 or CID-44229067 alone or combined with 5 μg/ml γ-tri-DAP. After 24hr, cytosolic and membrane subcellular fractions were isolated and analyzed (10 μg protein) by immunoblotting using antibodies specific for α-tubulin (cytosolic marker), pan-cadherin (plasma membrane marker), NOD1 (using anti-FLAG antibodies) and RIP2. Short (s.e.) versus long (l.e.) exposures of blots are presented for some results. All data are derived for a single blot. Intervening lanes were graphically excised (vertical line) for efficiency of presentation. See also Figures S1-S3 and S8-S12.
To examine whether CID-1088438 binds NOD1, we expressed and purified recombinant NOD1 protein from human cells and performed one-dimensional nuclear magnetic resonance (1D 1H-NMR) spectroscopy as a means to examine ligand binding. Indeed, the proton (1H) signal intensity derived from CID-1088438 was suppressed in the presence of NOD1 but not various control proteins such as Bcl-XL and Bid, thereby demonstrating direct interaction between this compound and NOD1 protein (Fig. 3B). However, the spectrum of the inactive analog compound CID-44229067 was also suppressed by NOD1 protein (not shown), which suggests that this compound may also bind NOD1 but fails to suppress its cellular activity. Similar results were obtained by affinity selection mass spectrometry (not shown). Interestingly, CID-1088438 did not interfere with ATP binding to recombinant NOD1 protein (Suppl. Fig. 8).
We obtained evidence that CID-1088438 may alter the conformation of NOD1 protein in vitro. For example, in experiments using purified His6-tagged NOD1, addition of active compound CID-1088438 but not inactive analog CID-44229067, markedly increased the relative amount of His6-NOD1 protein that bound to nickel-chelating resin (Ni/NTA) without affecting the binding of other His6-tagged control proteins such as Bcl-XL and Bid (Figure 3C). Similar results were obtained when CID-1088438 was added to living cells expressing His6-FLAG-NOD1 protein (Fig. 3D). In contrast, this compound had no effect on the efficiency that His6-myc-NOD2 protein was pulled down by Ni/NTA (Fig. 3D). Interestingly, NOD1 ligand γ-tri-DAP also impacted the efficiency with which His6-FLAG-NOD1 protein was pulled down with Ni/NTA, reducing the relative amount of NOD1 protein recovered from cells treated with either inactive or active compounds without changing total levels of His6-FLAG-NOD1 protein in lysates (Figure 3D). Addition of proteasome-inhibitor MG132 largely negated the effects of both γ-tri-DAP and compounds on His6-FLAG-NOD1 pull-down by Ni/NTA, suggesting a role also for ubiquitination in controlling NOD1 protein conformation. Indeed, we have obtained evidence that NOD1 undergoes both lysine 48 (K48) and K63-linked poly-ubiquitination (Suppl Fig. 9).
Next, we interrogated the effects of CID-1088438 on endogenous NOD1 protein. Because endogenous NOD1 is present at low levels and very difficult to detect with currently available antibodies, we enriched NOD1 protein from lysates of HCT-116 cells by immunoprecipitation, then analyzed NOD1 levels by immunoblotting. In unstimulated cells, no clear difference in the levels of NOD1 protein was observed following treatment with active compound CID-1088438 or inactive analog CID-44229067 (Fig. 3E). However, culturing cells with γ-tri-DAP resulted in a decline in NOD1 protein levels in cells treated with DMSO control or inactive compound but not active compound. Changes in NOD1 protein levels did not correlate with significant alterations in NOD1 (or NOD2 and RIP2) mRNA expression (Suppl. Fig. 5).
Upon activation by bacterial ligands, NOD1 is reported to oligomerize and form complexes with various cytosolic proteins including RIP2, SGT-1, IAPs, and Bid (da Silva Correia et al., 2007; Inohara et al., 1999; Krieg et al., 2009; Krieg and Reed, 2010; Yeretssian et al., 2011). However, CID-1088438 did not interfere with NOD1 association with RIP2, Bid, or SGT-1 under over-expressing conditions (Suppl. Figs. 10-11 and data not shown), suggesting that direct interference with protein-protein interactions unlikely explains the mechanism of this NOD1 inhibitory compound.
NOD1 is reported to traffic between membranes and cytosol (Kufer et al., 2008; Lecine et al., 2007), correlating with NF-κB activation (Kufer et al., 2008). We therefore performed subcellular fractionation analysis of MCF-7 cells stably expressing epitope-tagged NOD1 (Fig. 3F) or NOD2 (suppl. Fig. 12), which are conveniently detected by immunoblotting using epitope-specific antibodies. Cells were treated with our without the respective NOD1 or NOD2 activators (γ-tri-DAP or MDP), in the presence or absence of compounds. Remarkably, CID-1088438 induced enrichment of NOD1 protein in the membrane fraction, independently of γ-tri-DAP induction. No changes of NOD2 compartmentalization were observed upon treatment with CID-1088438 or CID-44229067 (suppl. Fig. 12). In contrast, CID-1088438 reduced membrane localization of RIP2, a NOD1-binding partner required for NF-κB induction (Fig. 4F, high exposure). These data are in concordance with reports that migration of RIP2 to the membrane is essential for stimulating NF-κB signaling (Kufer et al., 2008; Lecine et al., 2007). We conclude that CID-1088438 alters subcellular targeting of NOD1.
SIGNIFICANCE
We report the characterization of a new class of 2-aminobenzimidazole derivatives as the first selective NOD1 inhibitors. These compounds suppress NF-κB signaling induced by NOD1, but not by multiple other NF-κB activators tested, including NOD2, TLRs, TNF family cytokines, RNA Receptors (MAVS-activators), DNA damaging agents, and PKC activators. The active 2-aminobenzimidazoles also selectively interfere with the NOD1-mediated stimulation of stress kinases and the interferon response pathway. Our studies of the mechanisms of CID-1088438 (ML130), hereafter dubbed Noditinib-1, suggest that this compound modulates NOD1 conformation and alters its subcellular localization. As such, Noditinib-1 provides a useful research tool for interrogating the role of NOD1 in human cell specimens and a chemical probe for improving understanding of the complex mechanisms regulating NOD1 signaling functions. In this regard, pharmaceutical targeting of downstream components of NF-κB pathways results in pathway non-specific inhibition, causing broad suppression of host innate immune defenses and thus potentially leaving patients vulnerable to opportunistic infections. In contrast, compounds that selectively target upstream targets/pathways responsible for NF-κB signaling (such as NOD1) would be expected to leave most innate immunity defense mechanisms intact, but would be efficacious only for diseases where the specific target/pathway is causally involved in disease pathogenesis. Further elaboration of the mechanism of Noditinib compounds may reveal new strategies for achieving a proper balance of innate immunity responses and restoring health in conditions where NOD1 homeostasis is disrupted.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Hanaii, T. Siegfried, C. Wimer, M. Cuddy, M. Rossetti, H. Zhang, W. Yu (Sanford-Burnham Medical Research Institute, USA), P. Gosalia, C. Gasior, E. Suyama, S. Vasile, C. Hassig and T. Chung (Conrad Prebys Center for Chemical Genomics, CA, USA) for their assistance in this project. This work was supported by NIH Roadmap Initiative grant U54HG003916.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental information includes one table and twelve figures (with legends), as well as detailed experimental procedures.
Contributor Information
Ricardo G. Correa, Sanford-Burnham Medical Research Institute
Pasha M. Khan, Sanford-Burnham Medical Research Institute
Nadav Askari, Sanford-Burnham Medical Research Institute.
Dayong Zhai, Sanford-Burnham Medical Research Institute.
Motti Gerlic, Sanford-Burnham Medical Research Institute.
Roberto Spreafico, Sanford-Burnham Medical Research Institute.
Salvatore Albani, Sanford-Burnham Medical Research Institute.
Paul W. Diaz, Sanford-Burnham Medical Research Institute
Gregory P. Roth, Sanford-Burnham Medical Research Institute
REFERENCES
- Allison CC, Kufer TA, Kremmer E, Kaparakis M, Ferrero RL. Helicobacter pylori induces MAPK phosphorylation and AP-1 activation via a NOD1-dependent mechanism. J Immunol. 2009;183:8099–8109. doi: 10.4049/jimmunol.0900664. [DOI] [PubMed] [Google Scholar]
- Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GH, et al. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- Carneiro L, Magalhaes J, Tattoli I, Philpott D, Travassos L. Nod-like proteins in inflammation and disease. J Pathol. 2008;214:136–148. doi: 10.1002/path.2271. [DOI] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- da Silva Correia J, Miranda Y, Austin-Brown N, Hsu J, Mathison J, Xiang R, Zhou H, Li Q, Han J, Ulevitch RJ. Nod1-dependent control of tumor growth. Proc Natl Acad Sci U S A. 2006;103:1840–1845. doi: 10.1073/pnas.0509228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Correia J, Miranda Y, Leonard N, Ulevitch R. SGT1 is essential for Nod1 activation. Proc Natl Acad Sci U S A. 2007;104:6764–6769. doi: 10.1073/pnas.0610926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann L, Karin M. NOD2 and Crohn’s disease: loss or gain of function? Immunity. 2005;22:661–667. doi: 10.1016/j.immuni.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Franchi L, Park JH, Shaw MH, Marina-Garcia N, Chen G, Kim YG, Nunez G. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, Inohara N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. Embo J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Knoefel WT, Reed JC. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci U S A. 2009;106:14524–14529. doi: 10.1073/pnas.0907131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg A, Reed JC. IAPs and their emergent role in NLR signaling. Cell Cycle. 2010;9:426–427. doi: 10.4161/cc.9.3.10722. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008;10:477–486. doi: 10.1111/j.1462-5822.2007.01062.x. [DOI] [PubMed] [Google Scholar]
- Lecine P, Esmiol S, Metais JY, Nicoletti C, Nourry C, McDonald C, Nunez G, Hugot JP, Borg JP, Ollendorff V. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282:15197–15207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- Nishio H, Kanno S, Onoyama S, Ikeda K, Tanaka T, Kusuhara K, Fujimoto Y, Fukase K, Sueishi K, Hara T. Nod1 ligands induce site-specific vascular inflammation. Arterioscler Thromb Vasc Biol. 2011;31:1093–1099. doi: 10.1161/ATVBAHA.110.216325. [DOI] [PubMed] [Google Scholar]
- Opitz B, Puschel A, Beermann W, Hocke AC, Forster S, Schmeck B, van Laak V, Chakraborty T, Suttorp N, Hippenstiel S. Listeria monocytogenes activated p38 MAPK and induced IL-8 secretion in a nucleotide-binding oligomerization domain 1-dependent manner in endothelial cells. J Immunol. 2006;176:484–490. doi: 10.4049/jimmunol.176.1.484. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, Sassetti CM, Kelliher MA. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli V, Dostert C, Muruve DA, Tschopp J. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19:615–622. doi: 10.1016/j.coi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Rosenzweig HL, Galster KT, Planck SR, Rosenbaum JT. NOD1 expression in the eye and functional contribution to IL-1beta-dependent ocular inflammation in mice. Invest Ophthalmol Vis Sci. 2009;50:1746–1753. doi: 10.1167/iovs.08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. doi: 10.1038/sj.cr.7310019. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Barr MJ, Lukens JR, McGargill MA, Chi H, Mak TW, Kanneganti TD. Signaling via the RIP2 adaptor protein in central nervous system-infiltrating dendritic cells promotes inflammation and autoimmunity. Immunity. 2011;34:75–84. doi: 10.1016/j.immuni.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik C, Hayashi H, Pio F, Godzik A, Reed JC. CARD6 is a modulator of NF-kappa B activation by Nod1- and Cardiak-mediated pathways. J Biol Chem. 2003;278:31941–31949. doi: 10.1074/jbc.M300009200. [DOI] [PubMed] [Google Scholar]
- Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation. Semin Immunopathol. 2007;29:289–301. doi: 10.1007/s00281-007-0083-2. [DOI] [PubMed] [Google Scholar]
- Tergaonkar V. NFkappaB pathway: a good signaling paradigm and therapeutic target. Int J Biochem Cell Biol. 2006;38:1647–1653. doi: 10.1016/j.biocel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, Chiba T, Fuss IJ, Kitani A, Strober W. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. 2010;120:1645–1662. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeretssian G, Correa RG, Doiron K, Fitzgerald P, Dillon CP, Green DR, Reed JC, Saleh M. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 2011;474:96–99. doi: 10.1038/nature09982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.