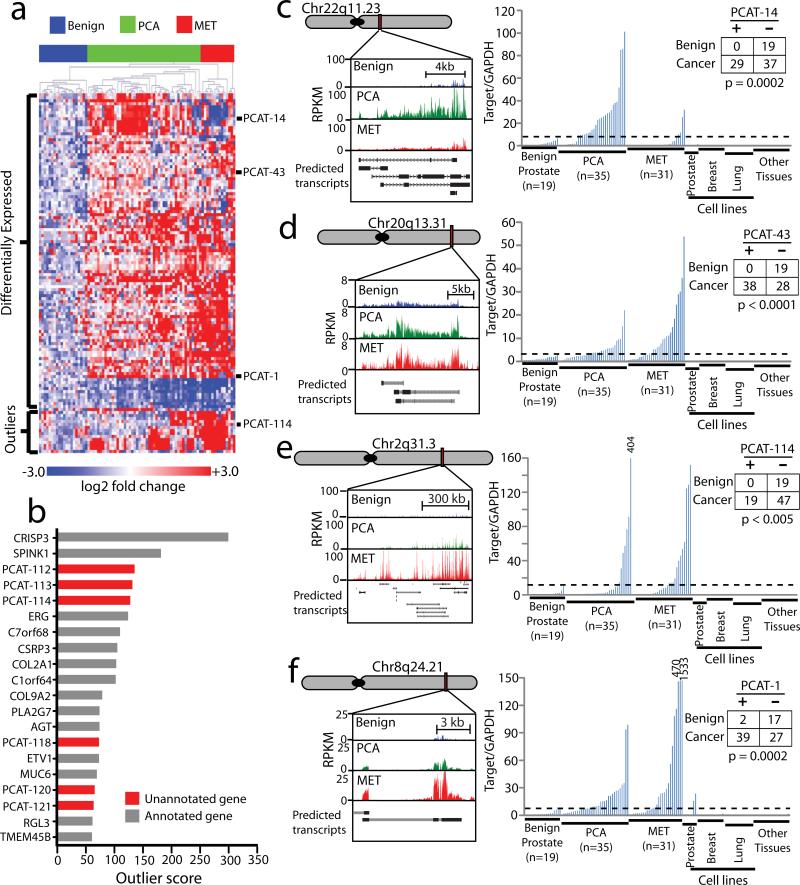
Figure 3. Unannotated intergenic transcripts differentiate prostate cancer and benign prostate samples.
(a) Unsupervised clustering analyses of differentially-expressed or outlier unannotated intergenic transcripts clusters benign samples, localized tumors, and metastatic cancers. Expression is plotted as log2 fold change relative to the median of the benign samples. The four transcripts detailed in this study are indicated on the side. (b) Cancer outlier expression analysis for the prostate cancer transcriptome ranks unannotated transcripts prominently. (c-f) qPCR on an independent cohort of prostate and non-prostate samples (Benign (n=19), PCA (n=35), MET (n=31), prostate cell lines (n=7), breast cell lines (n=14), lung cell lines (n=16), other normal samples (n=19), see Supplementary Table 8) measures expression levels of four nominated ncRNAs—PCAT-1, PCAT-43, PCAT-114, and PCAT-14—upregulated in prostate cancer. Inset tables on the right quantify “positive” and “negative” expressing samples using the cut-off value (shown as a black dotted line). Statistical significance was determined using a Fisher's exact test. (c) PCAT-14. (d) PCAT-43. (e) PCAT-114 (SChLAP1). (f) PCAT-1. qPCR analysis was performed by normalizing to GAPDH and the median expression of the benign samples.