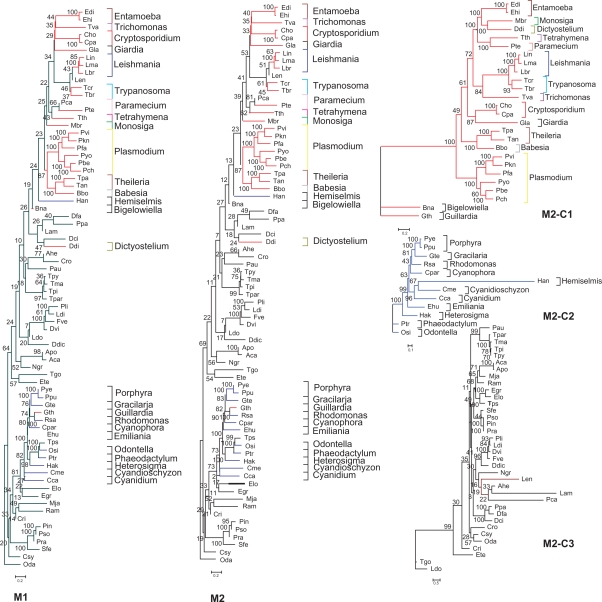
Figure 3.
Phylogenomic supermatrix trees of protozoan species using the total and trimmed (using TrimAl) alignments. Supermatrices of 21,260 (M1) and 12,807 (M2) positions were used respectively in 74 protozoan species. Maximum likelihood tree was constructed with Phyml 2.4.4, JTT as evolutionary model and bootstrap 100. The resulting clades of M2: M2-C1 in red, M2-C2 in blue and M2-C3 in black with the evolutionary models obtained with Modelgenerator 0.85 were used to construct three individual trees: M2-Ct1 (Blosum62), M2-Ct2 (RtREV) and M2-Ct3 (WAG).