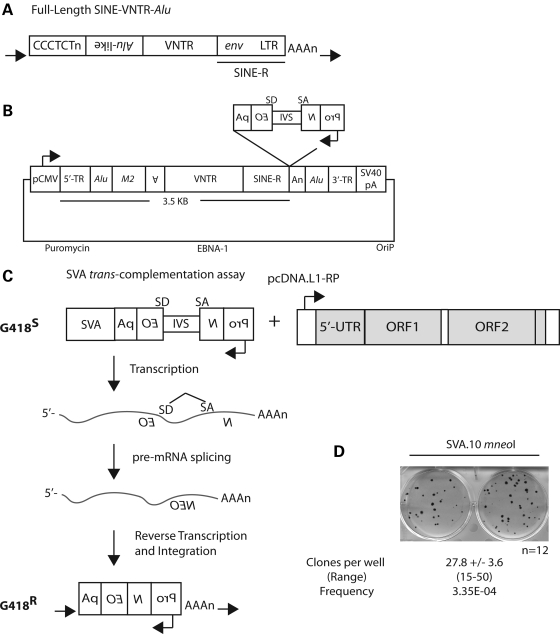
Figure 1.
A cell-culture SVA retrotransposition assay. (A) A full-length ‘canonical' SVA in the human genome, with the individual domains in order from 5′ to 3′. (i) CCCTCT hexamer; (ii) the Alu-like domain consisting of two antisense-spliced Alu fragments and a sequence of unknown origin; (iii) VNTR; and (iv) SINE-R (env sequence and right LTR from an extinct HERV-K), terminating in a polyA tail (AAAn), with the entire insertion flanked by a TSD (black horizontal arrows). (B) The SVA.10 mneoI construct. A ‘master' SVA locus, SVA.10, from the SVAF1 (MAST2) subfamily containing both 5′ (5′ TR, Alu) and 3′ (Alu, 3′ TR) transductions (TR) marked with the mneoI retrotransposition cassette cloned into the pCEP-Pur plasmid backbone. (C) The rationale of the trans-complementation assay is illustrated. Only if the SVA containing the mneoI reporter undergoes a round of transcription, followed by reverse transcription and integration presumably mediated by a full-length L1 (shown), will the reading frame of the neomycin phosphotransferase reporter be restored, conferring G418 resistance (G418R). (D) G418R foci formation is observed in HeLa HA cells when SVA.10 mneoI is co-transfected with the highly active L1 driver construct, pcDNA.L1-RP. The mean number of clones per well ± SEM and the range of clones across the wells are displayed below. The number of wells (n) assayed for this experiment is shown. The retrotransposition frequency (mean number of clones/number of transfected cells) for SVA.10 mneoI is listed below the range.