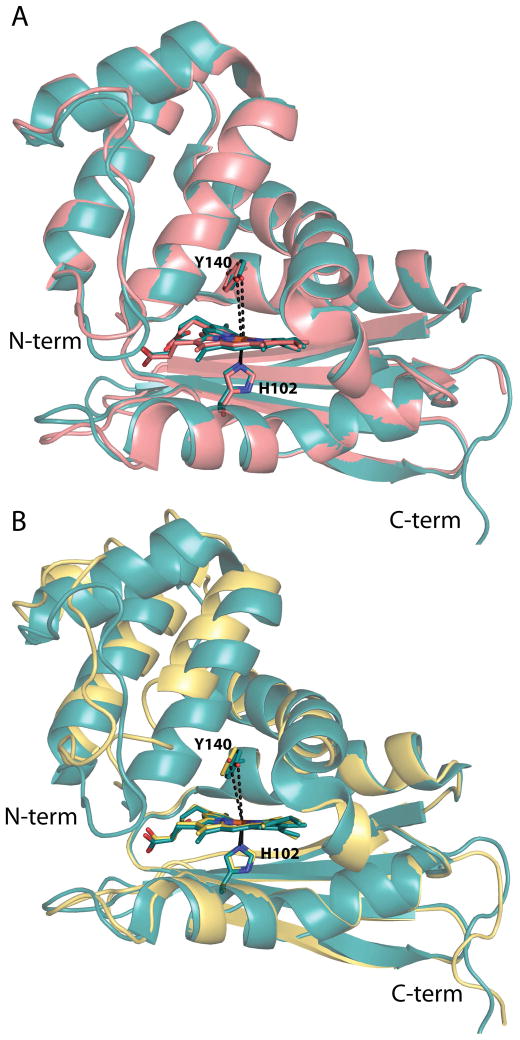
Figure 4.
Structural alignments of the WT Tt H-NOX structure with the I5L and I5F mutant structures. (A) Global alignment of WT (teal, PDB ID 1U55) (15) and I5L (salmon, PDB ID 3NVR) (17) Tt H-NOX structures; RMSD 0.34 Å. The Y140–Fe distances are shown in dashed black lines and are 5.0 and 5.1 Å in the WT and I5L structures, respectively. (B) Overlays of the WT (teal, PDB ID 1U55) (15) and I5F (yellow) mutant structures shown following alignment of the last 80 amino acids (C-terminus) of the proteins; RMSD 1.13 Å. The Y140–Fe distances are shown in black dashed lines with the I5F distance being 5.6 Å. H102, Y140, and the heme are shown in sticks for orientation.