Figure 1.
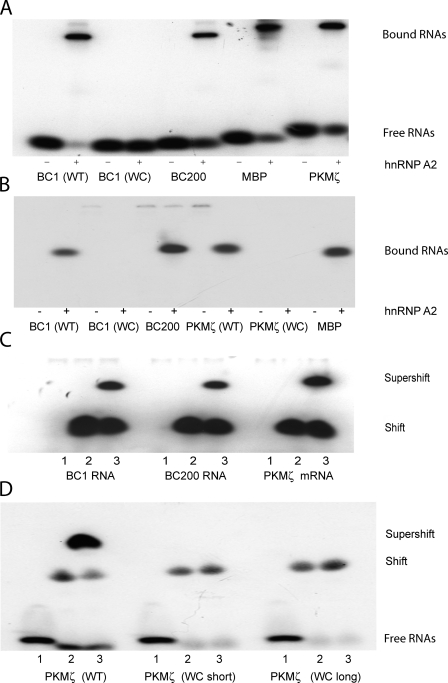
Dendritically transported BC1 RNA and PKMζ mRNA interact with hnRNP A2. EMSA experiments were performed using native PAGE. (A and B) BC1 RNA and PKMζ mRNA bind to recombinant hnRNP A2. EMSA experiments revealed shifts to lower mobility as a result of binding of radiolabeled RNAs to the protein. (A) Gel shifts induced by hnRNP A2 were observed with BC1 RNA, BC200 RNA, PKMζ mRNA, and MBP mRNA. For BC1 RNA and PKMζ mRNA, derivatives were used in which the noncanonical GA motif had been converted to canonical WC base pairing. Such WC mutants, in contrast to the respective WT species, failed to interact with hnRNP A2 (A and B; only bound RNAs are shown in B). For secondary structures of WT and WC mutant motifs in BC1 RNA and PKMζ mRNA, see Fig. S1. The PKMζ mRNA WC mutant used in B was variant long. (C and D) BC1 RNA, BC200 RNA, and PKMζ mRNA are recognized by hnRNP A2 in brain extracts. (C) An antibody specific for hnRNP A2 (Ma et al., 2002) produced a supershift with BC1 RNA, BC200 RNA, and PKMζ mRNA. Shifted and supershifted bands are shown. (D) Recognition of PKMζ mRNA by hnRNP A2 in brain extracts depends on a noncanonical motif structure in a 3′ UTR stem loop. WT PKMζ mRNA was supershifted by antibodies against hnRNP A2; in contrast, WC mutant PKMζ mRNAs were not. Loading (C and D): 1, RNAs; 2, RNAs plus brain extract; 3, RNAs plus brain extract plus anti-A2 antibody.