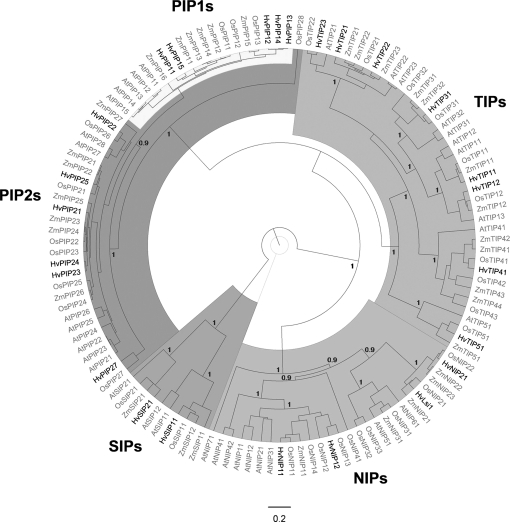
Fig. 2.
Bayesian phylogenetic analysis of barley major intrinsic proteins (MIPs). Barley MIP protein sequences were aligned with MIPs of Arabidopsis, maize, and rice using ClustalW. Barley MIPs analysed are highlighted. Annotations and protein sequences are given in Supplementary File S2 at JXB online. Concerning HvNIP2;1, Fig. 2 refers to the gene annotated first (accession number BA166444) and not to the gene referred to by Schnurbusch et al. (2010) as a boron transporter (HvNIP2;1). The latter gene is identical in sequence to the silicon transporter (HvLsi1, shown) reported by Chiba et al. (2009). HvSIP2;1 is not annotated; only a partial sequence is known, which shows the highest homology to AtSIP2;1 among Arabidopsis SIPs. The distance scale represents the evolutionary distance, expressed as the number of substitutions per amino acid. The figures displayed on the main nodes reflect the posterior probability.