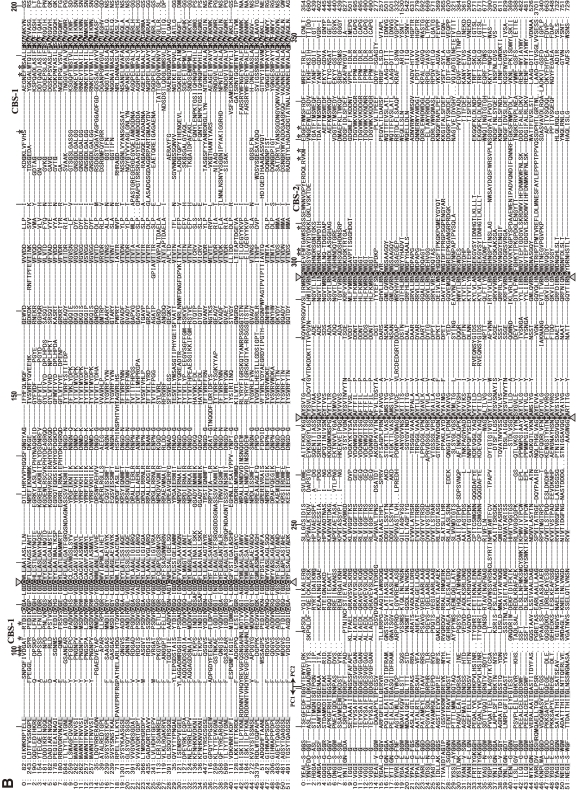
Figure 8.
Phylogenetic tree and sequence alignment of bacterial calpains. A. Phylogenetic tree of bacterial calpains and human CAPN1/μCL. See Fig. 3A for methods. B. Sequence alignment of the bacterial calpain protease domains and human CAPN1/μCL. Only the sequences of the protease domains are compared, with the start and end aa residue numbers indicated, as other sequences and lengths are somewhat divergent. Reversed fonts and gray shadow indicate residues conserved in all sequences or more than half, respectively. Triangles: active site residues. Asterisks: residues involved in Ca2+-binding of the protease domain of human CAPN1/μCL. Sequences: 0: H. sapiens NP_001185798; 1: Acaryochloris marina MBIC11017 YP_001520592; 2: Actinomyces odontolyticus ATCC 17982 ZP_02043925; 3: Actinomyces odontolyticus F0309 ZP_06609922; 4: Actinomyces sp. oral taxon 848 str. F0332 ZP_06161744; 5: Actinomyces sp. oral taxon 848 str. F0332 ZP_06161892; 6: Actinomyces sp. oral taxon 848 str. F0332 ZP_06162803; 7: Actinomyces sp. oral taxon 848 str. F0332 ZP_06162809; 8: Anabaena variabilis ATCC 29413 YP_322403; 9: Bacteroides ovatus ATCC 8483 ZP_02064250; 10: Bacteroides sp. 1_1_14 ZP_06994459; 11: Bacteroides sp. 1_1_6 ZP_04847694; 12: Bacteroides sp. D2 ZP_05758284; 13: Bacteroides thetaiotaomicron VPI-5482 NP_812871; 14: Brachybacterium faecium DSM 4810 YP_003153779; 15: Brachybacterium faecium DSM 4810 YP_003155356; 16: Bradyrhizobium sp. BTAi1 YP_001241302; 17: Bradyrhizobium sp. ORS278 YP_001204811; 18: Brevibacterium mcbrellneri ATCC 49030 ZP_06806606; 19: Cyanothece sp. PCC 7822 YP_003887732; 20: Cyanothece sp. PCC 7822 YP_003887741; 21: Frankia alni ACN14a YP_716900; 22: Frankia sp. CcI3 YP_483525; 23: Frankia sp. EAN1pec YP_001504490; 24: Frankia sp. EuI1c YP_004014028; 25: Frankia sp. EUN1f ZP_06411021; 26: Gemmata obscuriglobus UQM 2246 ZP_02733073; 27: Geobacter lovleyi SZ YP_001952615; 28: Gloeobacter violaceus PCC 7421 NP_927086; 29: Granulibacter bethesdensis CGDNIH1 YP_745367; 30: Herpetosiphon aurantiacus ATCC 23779 YP_001544076; 31: Herpetosiphon aurantiacus ATCC 23779 YP_001545619; 32: Herpetosiphon aurantiacus ATCC 23779 YP_001546212; 33: Legionella longbeachae D-4968 ZP_06188344; 34: Legionella longbeachae NSW150 YP_003455673; 35: Microcystis aeruginosa NIES-843 YP_001659697; 36: NC10 bacterium ‘Dutch sediment’ CBE68371; 37: Nostoc sp. PCC 7120 NP_484413; 38: Nostoc sp. PCC 7120 NP_485127; 39: Nostoc sp. PCC 7120 NP_487855; 40: Photorhabdus luminescens subsp. laumondii TTO1 NP_929692; 41: Planctomyces limnophilus DSM 3776 YP_003628665; 42: Porphyromonas gingivalis AAA25652; 43: Porphyromonas gingivalis W83 NP_905271; 44: Rhodopseudomonas palustris BisA53 YP_780017; 45: Salinispora tropica CNB-440 YP_001157346; 46: Segniliparus rotundus DSM 44985 YP_003659885; 47: Streptomyces albus J1074 ZP_06592718; 48: Streptomyces viridochromogenes DSM 40736 ZP_07305717; 49: Synechococcus sp. RS9916 ZP_01472825; 50: Synechococcus sp. RS9916 ZP_01472826; 51: Synechococcus sp. RS9916 ZP_01472827.