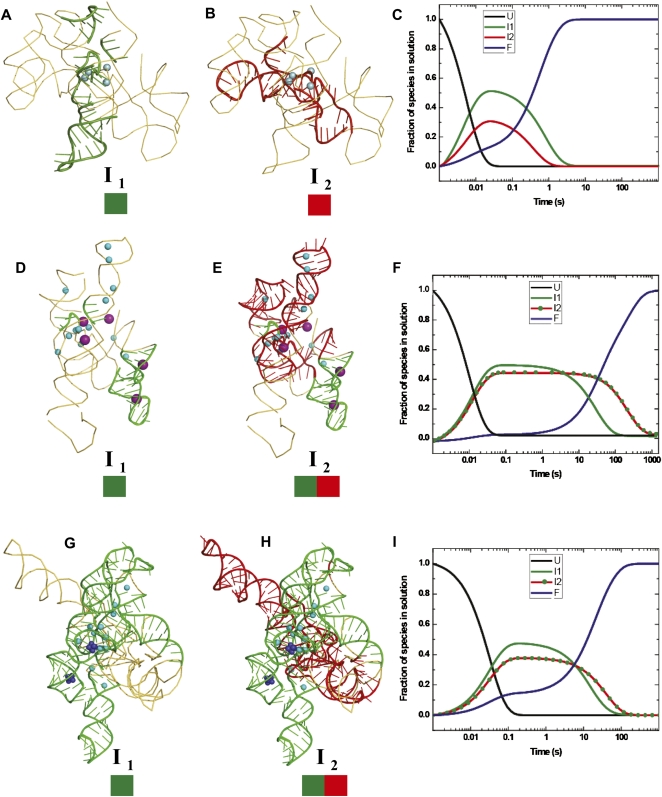
FIGURE 4.
Mapping the temporal clusters of tertiary interactions onto the crystal structures of the Twort (A,B) and Azoarcus (D,E) ribozymes, and the biochemically validated phylogenetic model of the Tetrahymena (G,H) ribozyme. The coordinates of the structures were obtained from the following PDB entries: 1Y0Q (Twort), 1ZZN (Azoarcus), and TtLSU (Tetrahymena). The positions of divalent and monovalent ions (when known in the crystal structures) are indicated by blue and purple spheres, respectively. The green and red colored boxes drawn below the intermediates (I1 and I2) represent the clusters of tertiary contacts formed in each species. The schematic models of the intermediates were generated in PyMOL (DeLano 2009). The three right panels show the lifetimes in solution of the unfolded (U: black line), intermediate (I1: green line, I2: red line or red line with green dots), and the folded (F: blue line) species, as derived from KinFold analysis, for the Twort (C), Azoarcus (F), and Tetrahymena (I) ribozymes. The two deep blue clusters of spheres in the P4–P6 domain of the Tetrahymena ribozyme represent the two cobalt hexamine(III) molecules observed in the crystal structure (1GID.pdb).