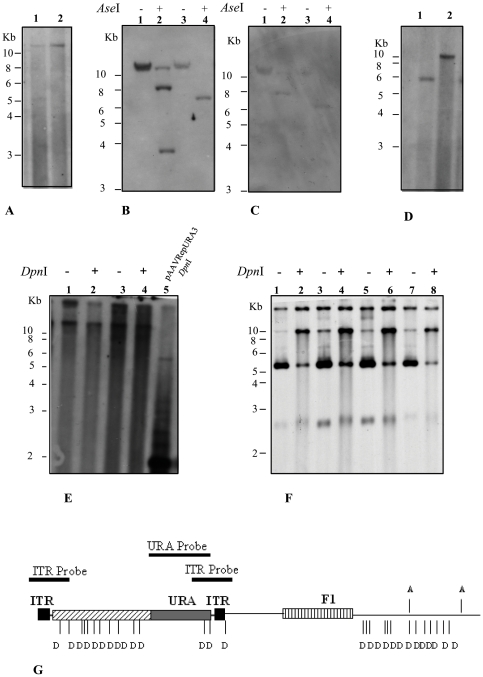
Figure 4. AAV replication in yeast.
(A) Southern blot analysis of low Mr DNA of two different yeast clones URA3+ (lane 1 and 2) derived from transformation with pAAVRepURA3 using the URA3 marker gene as probe. (B, C) Southern blot analysis of genomic DNA of the same two clones as in B undigested (lane 1 and 3), and digested with AseI (lane 2 and 4) probed with URA3 (B) or ITR probe (C). (D) Low Mr DNA of the two yeast clones URA3+LEU2+ derived from transformation with pAAVRepURA3 and successively transformed with pG.Rep68 (lane 1 and 2) analyzed on Southern Blot using the URA3 probe. (E) Low Mr DNA of yeast clones URA3+LEU2+ derived from co-transformation of pAAVRepURA with control plasmid pGAD424. Lanes 1 and 3 show the undigested DNA and lanes 2 and 4, DNA digested with DpnI and subjected to Southern blot analysis using URA3 marker gene as probe. Lane 5 shows the result of DpnI digestion of the pAAVRepURA plasmid. DpnI was performed in order to demonstrate that AAV DNA replicated in yeast. (F) Low Mr DNA of yeast clones URA3+LEU2+ derived from co-transformation of pAAVRepURA with plasmid pG.Rep68. Lanes 1, 3, 5, 7 show the undigested DNA and lanes 2, 4, 6, 8 DNA digested with DpnI and subjected to Southern blot analysis using URA3 marker gene as probe. (G) Schematic representation of DpnI/MboI (indicated with D) and AseI (indicated with A) restriction map of pAAVRepURA plasmid. The DpnI/MboI restriction endonucleases do not cut in the URA3 gene.