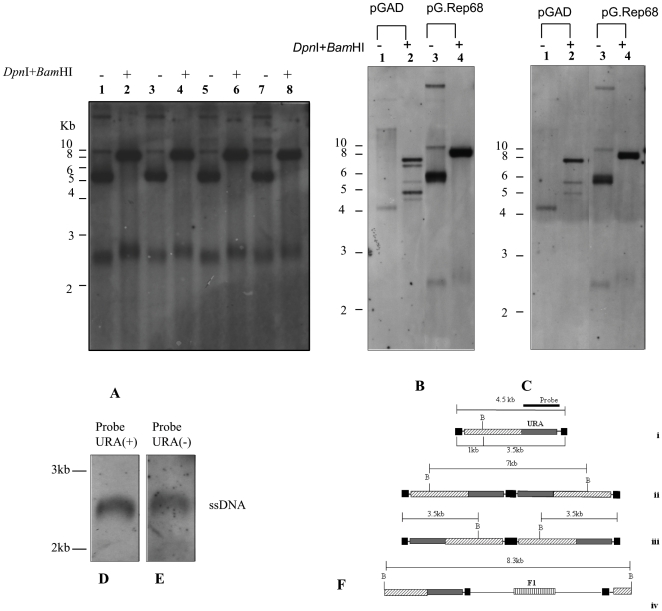
Figure 6. Characterization of newly replicated DNA.
(A) Low Mr DNA of four clones derived from yeast co-transformed with pAAVRepURA and pG.Rep68 digested with DpnI and BamHI (lane 2, 4, 6, 8) and not digested (lane 1, 3, 5, 7) was analyzed on Southern Blot and probe with the URA3 gene. BamHI cuts once in AAV construct. (B, C) Low Mr DNA of one clone obtained from the co-transformation of pAAVRepURA with pGAD not digested (lane 1) and digested with DpnI and BamHI (lane 2); the low Mr DNA of one out four clones analyzed in panel A undigested (lane 3) and digested with DpnI and BamHI (lane 4) was analyzed on Southern Blot and detected with URA3 probe (B) and F1 probe (C). (D, E) Equal amount of low Mr DNA of one out of four clones analyzed in panel A was loaded in two gel slots and transferred on nylon membrane. The membrane was cut in two pieces corresponding to gel slots and further cut to leave only ssDNA. The ssDNA was detected using as probe a 100-mer oligonucleotide complementary to the filament (+) of the URA gene URA(+) (D), or a oligonucleotide probe complementary to filament (-) of the URA gene URA(-) (E), to determine the polarity of ssDNA. (F) Restriction maps of predicted replicative intermediates: i) linear monomer, ii) tail-to-tail dimer, iii) head-to-head dimer, iv) linear vector. Position of BamHI is indicated with B. The sizes of the fragments liberated following BamHI cleavage and recognized by the URA probe are shown next to corresponding structures. ITRs are indicated as black boxes.