SUMMARY
Inositol 1,4,5-trisphosphate receptors (IP3Rs) play a fundamental role in generating Ca2+ signals that trigger many cellular processes in virtually all eukaryotic cells. So far, the three-dimensional (3D) structure of these channels has remained extremely controversial. Here, we report a subnanometer resolution electron cryomicroscopy (cryo-EM) structure of a fully functional type 1 IP3R from cerebellum in the closed state. The transmembrane region reveals a twisted bundle of four α-helices, one from each subunit, that form a funnel shaped structure around the 4-fold symmetry axis, strikingly similar to the ion-conduction pore of K+ channels. The lumenal face of IP3R1 has prominent densities that surround the pore entrance and similar to the highly structured turrets of Kir channels. 3D statistical analysis of the cryo-EM density map identifies high variance in the cytoplasmic region. This structural variation could be attributed to genuine structural flexibility of IP3R1.
INTRODUCTION
IP3Rs form a family of intracellular Ca2+ channels that integrate multiple cellular stimuli (including neurotransmitters, hormones, growth factors, odorants and light) to release Ca2+ from intracellular stores such as the endoplasmic reticulum (ER) into the cytoplasm (Foskett et al., 2007; Mikoshiba, 2007). IP3R channels are activated by Ca2+ and IP3, their primary ligands, and are modulated via complex interactions with numerous intracellular ligands. Such complex functionality implies multiple interactions and regulatory motifs with distinctive features that enable conversion of cellular signals into Ca2+ signals. Three types of IP3Rs (type 1–3) have been cloned in mammals and share ~70% sequence identity. Each type shows distinct properties in terms of modulation by endogenous and exogenous ligands. A functional IP3R channel is a homo- or hetero-tetrameric integral membrane complex of over 1.2 MDa, which in vivo directly associates with an array of accessory/modulatory proteins. Among the mammalian isoforms of IP3R, IP3R1 is the best characterized and the predominant type in cerebellar ER where it forms homotetrameric assemblies. Each subunit of IP3R1 is a 2749 residue protein that can be structurally divided into three regions: a large N-terminal region with an IP3-binding region close to the N-terminus; a trasmembrane (TM) region (residues 2275 – 2593) containing putative 6 membrane-spanning segments near the C-terminus, and a C-terminal tail (the last ~160 residues) (Furuichi et al., 1989; Mignery et al., 1989). A central mechanistic question in IP3R gating is how IP3 binding in the N-terminal sequence of the channel protein is communicated to the ion conduction pore formed near the C-terminus. Recently, the crystal structures of three domains comprising the N-terminal ligand-binding region were determined: the IP3-binding core region (residues 224–604) (Bosanac et al., 2002) and the suppressor domain (residues 2–223) (Bosanac et al., 2005). The structure of the IP3-binding core region consists of two domains, an N-terminal β-trefoil and a C-terminal armadillo repeat that forms a single IP3-binding site at their interface. The suppressor domain conforms to a β-trefoil and a helix-turn-helix motif shaped together as a hammer-like structure. Although this study provides valuable insights into spatial relationship between IP3-binding site and two putative Ca2+ binding sites, the spatial relationship between the suppressor and ligand-binding domains remains unknown. Despite accumulating biochemical, mutagenesis and functional studies (Boehning and Joseph, 2000; Schug and Joseph, 2006; Uchida et al., 2003) suggesting structural coupling between the N-terminal domains and the channel pore region, the structural basis for IP3-induced gating remains poorly understood due to the lack of knowledge about 3D architecture of the full-length channel protein.
Several low-resolution 3D structures of the full-length IP3R1 channel in the closed state have been previously reported based on single particle electron microscopy analysis (da Fonseca et al., 2003; Hamada et al., 2003; Jiang et al., 2002a; Sato et al., 2004; Serysheva et al., 2003). These structures are highly inconsistent, even at the coarsest structural level, leading to considerable uncertainty over its true 3D architecture. A common denominator exists among all these studies, which is an unusually high-level of noise in the raw cryo-EM images. Insufficient contrast can lead to erroneous reconstructions with either missing or artifactual features due to model bias (Stewart and Grigorieff, 2004). Through extensive efforts to optimize the biochemical purification and vitrification process, we have now produced cryo-EM images with dramatically higher contrast. This has permitted us to reconstruct the full channel, unambiguously, to roughly 1 nm resolution and to rationalize its biological functions in terms of its molecular architecture.
RESULTS and DISCUSSION
Structure Determination
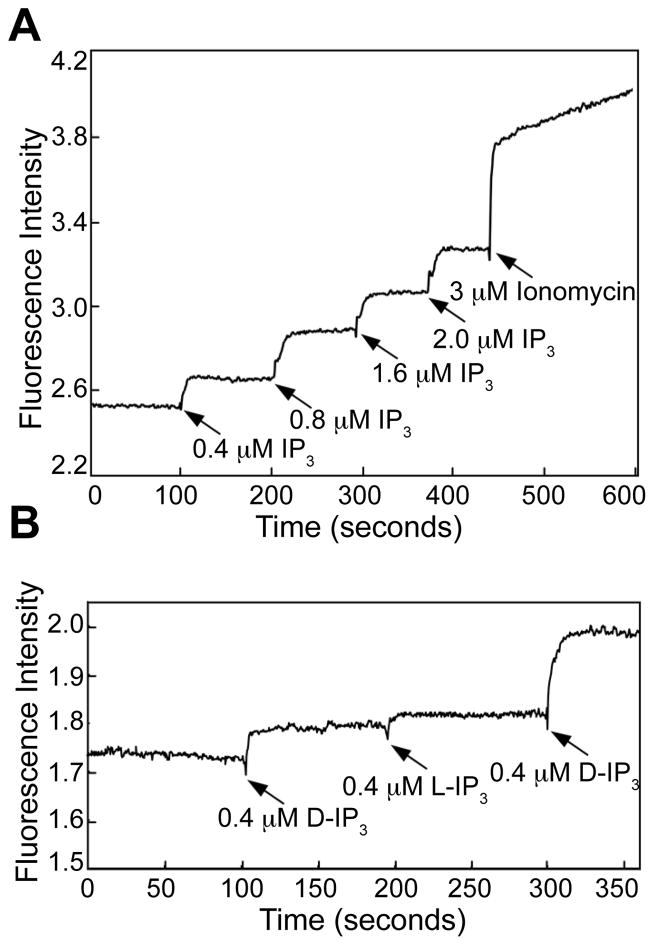
To perform structural analysis by single-particle cryo-EM reconstruction, IP3R1 channel was purified from rat cerebellum as previously described (Serysheva et al., 2003) with additional optimizations (Figure 1SA). To confirm that purified channels are functionally intact, we reconstituted them into Ca2+-loaded liposomes prior to cryo-EM imaging. IP3-triggered Ca2+ efflux experiments were performed and clearly demonstrate that the purified channels remain functional and exhibit ion-conducting properties characteristic of native IP3R1 channels (Figure 1 and Figure S1B) (Hirata et al., 1994; Marchant and Taylor, 1998).
Figure 1. Functional characterization of the purified IP3R1.
(A) IP3-induced activation of the purified IP3R1 reconstituted into lipid vesicles. Time-based Ca2+ efflux from IP3R1- liposomes was monitored by measuring the change in Fura-2 fluorescence. Stepwise addition of D-IP3 induced Ca2+ release from the IP3R1- liposomes up to ~80% from maximum Ca2+ available for efflux in the presence of 3 μM ionomycin. (B) Stereo-specific response of the purified IP3R1 to activation with the D-IP3 but not to the L-IP3 confirms pharmacological identity of the purified channel. The slight increase in Ca2+ efflux observed with addition of L-IP3 is due to the presence of contaminating D-IP3 in the commercially obtained L-IP3 (per communication with Sigma-Aldrich) and contaminating Ca2+ in the solution (see also Figure S1).
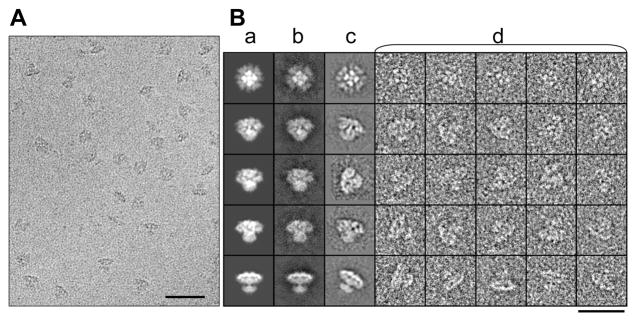
Electron images of ice-embedded IP3R1 (Figure 2A) were captured without adding any of its primary agonists, i.e. IP3 and Ca2+. These conditions favor the closed state of the channel (Hirata et al., 1994; Marchant and Taylor, 1998). Unlike previously published data, the cryo-EM data used in the present study has sufficient contrast that the shape of individual particles is clearly visible in the raw images (Figure 2 and Figure S2B). The relatively high signal/noise ratio in these images permitted reliable particle alignment during single particle reconstruction. The clear agreement between raw cryo-EM images, reference free class-averages, reference-based class-averages and reprojections of the 3D reconstruction is strong evidence for the accuracy of this map (Figure 2B). This reconstruction has been repeated using different starting models as well as different methods, and reliably produces a consistent structure. The resulting reconstruction is shown in Figure 3, Movie S1 and has a resolution of ~9.5 Å (Figure S2C, D).
Figure 2. Cryo-EM reconstruction of IP3R1.
(A) 200 kV image of IP3R1 embedded in a vitreous ice. The scale bar is 500 Å. (B) Projections (a) from the 3D reconstruction, corresponding class-averages generated in the final refinement loop (b), selected reference-free class-averages generated by 2D classification without a 3D reference (c), and a selection of corresponding unaligned particles (d). In our preparation, particle orientation was relatively uniform, meaning we expect resolution to be fairly isotropic. Scale bar is 400 Å (see also Figure S2).
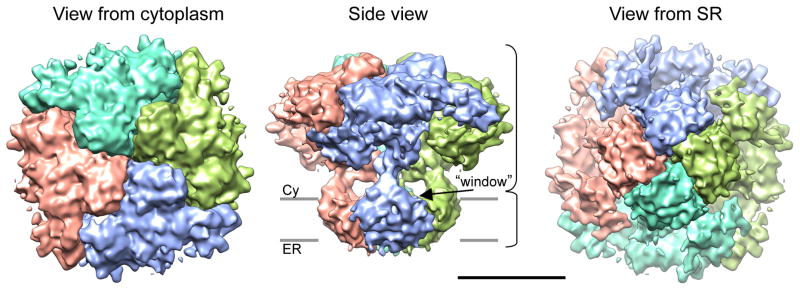
Figure 3. Surface representation of a cryo-EM density map of IP3R1 channel in the closed state.
The 3D structure of RyR1 is shown in three orthogonal views: a top view as seen from the cytoplasmic side, a side view and a bottom view as seen from the lumenal side. Subunits identified in the 3D structure of the channel tetramer are individually colored. The contour level corresponds to a protein mass of ~330 kDa per subunit at protein density of 0.81 Da/Å3. The grey bars suggest the positions of the cytoplasmic (Cy) and endoplasmic (ER) leaflets of the membrane bilayer. Scale bar is 100 Å (see also Figure S3 and Movie S1).
Overall Architecture of IP3R1
The IP3R1 channel adopts a mushroom-like shape that contains two square-shaped regions with side dimensions of ~ 220 Å and ~120 Å connected via thin stalk densities, together forming a ~190 Å tall structure (Figure 3, Figure S3, Movie S1).
Hydropathy analysis predicts that the C-terminal region of IP3R1 contains six TM helices between residues 2275–2593 suggesting that the TM region accounts for ~11% of the protein sequence, corresponding to ~36 kDa. Consistent with this membrane topology, volumetric estimation of our cryo-EM structure shows that the large square-shaped region and the stalk densities form a large cytoplasmic (CY) region comprising ~89% of the protein mass and include both the N- and C-termini of IP3R1. The volume of the TM region observed in the current 3D reconstruction is ~59,000 Å3, ~15% of the total volume of the channel subunit and accounts for a mass of ~ 48 kDa, assuming the protein density is 0.81 Da/Å3. This value is in reasonable agreement with the predicted 6-TM topology of IP3R1 protein, particularly when the presence of an unknown amount of detergent and residual lipid bound to the periphery of the TM domains is accounted for. Unfortunately, it is impossible to distinguish the boundary between protein and detergent or residual lipid in the TM domain, so it is impossible to conclusively determine the amount of bound detergent from our structure. In addition, the luminal portion of the IP3R1 channel is glycosylated, which would contribute some mass corresponding to carbohydrate moieties (Michikawa et al., 1994).
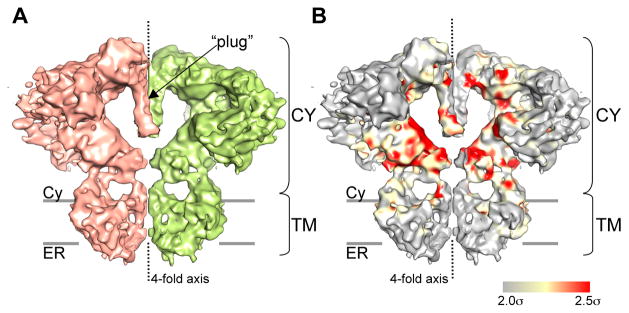
While the overall architecture of IP3R1 is somewhat similar to that of RyR channels, there are notable differences at a detailed structural level (Figure 4A and Figure S4). The CY region of IP3R1 includes a complex pattern of densities and large water-filled regions, also present in RyRs. Four IP3R1 subunits make extensive inter-subunit contacts via a central ‘plug‘ along the 4-fold channel axis (Figure 4A and Movie 1). This region appears to be a primary contact holding the four subunits together, and is completely absent in RyR channels (Figure S4). The CY region communicates with the TM region via stalk densities of the column region. The stalk densities shape the four side ‘windows’ located directly over the TM region at the subunit interfaces (Figure 3 and Figure S3). These ‘windows’ are conceptually similar to the lateral openings in K+ channels (Doyle et al., 1998; Kuo et al., 2003; Tao et al., 2009) and in the nicotinic acetylcholine receptor (nAchR) (Unwin, 2005), and may allow Ca2+ ions to flow freely between the channel domains and the cytoplasm. However, uncertainties in isosurface at this resolution make any detailed assessments of size impossible.
Figure 4. The internal architecture of IP3R1.
(A) Two opposing subunits are shown in a side view. (B) The 3D statistical variance map of the IP3R1 structure. The surfaces of individual IP3R1 subunits are colored according to the variance map. The threshold for color-coding is set to 2.5σ over the mean density in the 3D variance map. Regions of the highest variance are shown in red, the grey regions show no significant variations. Higher variances are observed in the core of the CY region, with the solvent exposed ‘scaffolding’ of the CY region and the TM region being more rigid. The grey bars indicate the putative position of the membrane bilayer (see also Figure S4 and Movie S2).
The present 3D structure of IP3R1 differs from the earlier cryo-EM reconstructions of IP3R1 (Jiang et al., 2002a; Sato et al., 2004; Serysheva et al., 2003), but it is consistent with the higher contrast(but much lower resolution) 3D structures of IP3R1 based on negative staining EM (da Fonseca et al., 2003; Hamada et al., 2003). It is impossible to prove the true source of the discrepancies among the previously published cryo-EM structures (Jiang et al., 2002a; Sato et al., 2004; Serysheva et al., 2003), and we cannot rule out the possibility of previous problems with biochemical purification or functional state of the channel as contributing factors (Table S1). High noise levels in the raw data, our own previously published data included, are by far the most likely explanation.
Statistical Validation of Cryo-EM Density Map
To better assess the structure and to validate the resolved structural features, we also analyzed the variance of the 3D structure (Chen et al., 2008; Penczek et al., 2006). A 3D standard deviation was estimated from 100 3D reconstructions produced using random resampling of particles with replacement applied to individual class-averages. This variance map shows the highest variability is in a portion of the CY region (Figure 4B), which exhibits a relatively rigid surface with a more flexible interior. In addition, in 2-D class averages, significant variations in the corresponding part of the CY region, and some motion of the TM region with respect to the CY region can be observed (Movie S2). It is likely that the unliganded closed state will thus be more conformationally flexible than the ligand-bound state. This observation suggests the existence of multiple conformations of the CY domains within this closed state maintained in the absence of channel primary ligands, i.e. IP3 and Ca2+. Our observations on a full-length protein are consistent with NMR and small-angle X-ray scattering studies performed with the IP3-binding and suppressor domains expressed as independent entities (Chan et al., 2007). This study shows that the N-terminal region exhibits a less compact structure in IP3-free state resulting from flexibility in the linker between IP3-binding domains. In addition, conformational variability of the CY region observed in our cryo-EM structure could be also attributed to the presence of unknown contaminating ligands co-purified with the IP3R1 protein, subtly altering its conformation.
The Pore Structure
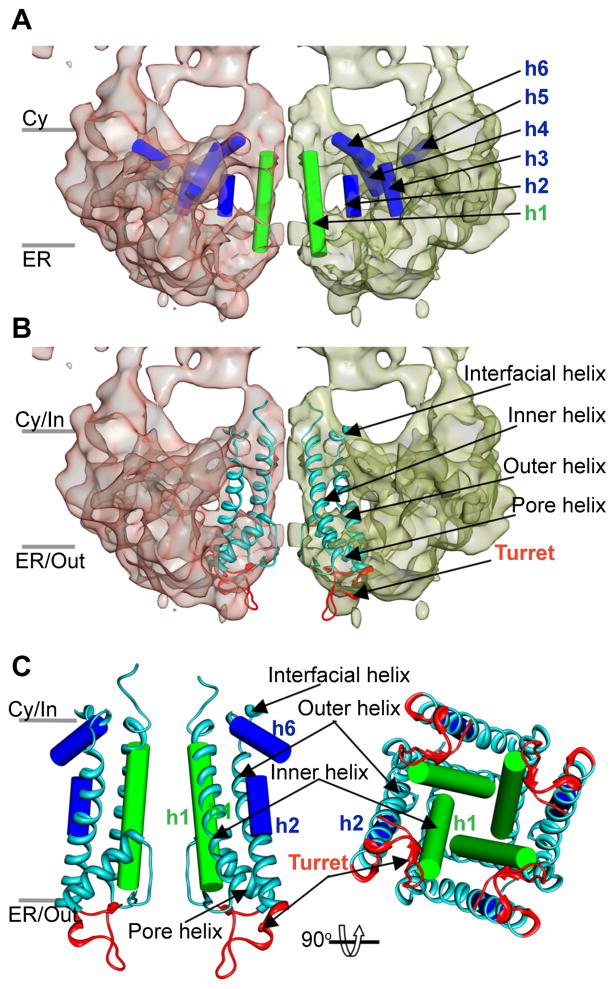
The lower variance of the TM region permitted us to search for the long α-helices which emerge at ~1 nm resolution (Baker et al., 2007; Baker et al., 2010). Six putative α-helices were identified in the TM region (Figure 5A and Figure S5A, Movie S1). Helix 1 lies near the central channel axis, and spans a length of ~26 Å, within the thickness of the hydrophobic core of the membrane. Four such rod-shaped densities (one from each subunit) form a twisted bundle arranged as a funnel with the wider end on the lumenal side. The helix 1 lining pathway thus narrows from ~28 Å at the lumenal vestibule to ~14 Å closer to the cytoplasmic side (Figure 5). These approximate dimensions are center-to-center separations between two opposing helices and do not account for amino acid side chains, which clearly cannot be resolved at the current resolution. This packing is strikingly similar to that of the inner, pore-lining α-helices in closed-state K+ channels (Figure 5B, C; Movie S1) (Doyle et al., 1998; Kuo et al., 2003; Tao et al., 2009), suggesting that helix 1 lines the ion-conduction pathway in the pore of the IP3R1 channel and that the region of minimum separation between the four helix 1’s represents the location of the channel gate.
Figure 5. Close-up view of the TM region.
(A) Putative secondary structure elements identified in the TM structure of IP3R1. TM regions of two opposing IP3R1 subunits are viewed from the side parallel to the membrane. The densities attributed to α-helices identified are annotated with cylinders and arbitrarily labeled h1–6 for reference. (B) X-ray structure of the eukaryotic Kir2.2 channel (PDB ID: 3JYC) is shown as a ribbon fitted into the cryo-EM density map of IP3R1; structural elements of the Kir channel pore are indicated. (C) Secondary structure elements identified in the pore region of IP3R1 are superimposed with the X-ray structure of the Kir2.2 channel: left panel - two subunits are viewed parallel to the membrane plane; right panel - four subunits are viewed along the channel 4-fold axis from the lumenal side, the pore helices of Kir2.2 channel are removed for clarity. The grey bars mark the putative positions of the Cy/In and ER/Out leaflets of the membrane bilayer (see also Figure S5 and Movie S1).
Short helices 2, 3 and 4 (Figure 5A and Figure S5A) are not long enough to span the lipid bilayer, but may represent a segment of longer helices not entirely resolved in this map. Based on their orientations with respect to the membrane plane, these rod densities may represent three other predicted TM helices. The position of helix 2 with respect to the inner helix found in our current structure of IP3R1 is similar to that of the outer helix in the K+ channels (Figure 5B, C; Movie S1) (Doyle et al., 1998; Kuo et al., 2003; Tao et al., 2009). Two other α-helices (labeled 5 and 6) run almost parallel to the membrane plane at the interface between the TM and CY regions (Figure 5A). Helix 6 is similarly placed with the interfacial helix of RyR1 (Ludtke et al., 2005) and Kir channels (Figure 5B, C; Movie S1) (Kuo et al., 2003; Tao et al., 2009). While the putative helices we have identified are only those, which were identified independently by both a human, and SSEhunter, at the current resolution, these helices remain somewhat speculative, and they will likely be refined as resolution improves.
Sequence Assignment for the Pore Region
Primary sequences of IP3R1 and RyR1 channels share high sequence homology in the C-terminal region encompassing the last two predicted TM helices (Figure S5B). This suggests common structural elements in the channel region and some similarity with aspects to channel function. Based on amino acid sequence alignments and guided by crystal structures of K+ channels (Doyle et al., 1998; Kuo et al., 2003; Tao et al., 2009), we propose that the predicted TM6 sequence (residues 2570–2593) correlates with the inner helix (Figure 5 and Figure S5B, C). This sequence would correspond to helix 1 identified in this study. Our sequence assignment for the inner pore-lining helix places the IP3R1 hinge region with the conserved Gly2587 (Figure S5B) within the membrane bilayer that permits bending of the inner TM helix during channel gating, similar to that in MthK channel (Jiang et al., 2002b). Moreover, this positions two hydrophobic residues Phe2593 and Ala2594 (Figure S5B) about the cytoplasmic vestibule in the present pore structure. The activation gate is a favorable position for the large hydrophobic residues as they act to constrict the pore entryway, as it was demonstrated for nAchR (Miyazawa et al., 2003), MscL (Chang et al., 1998) and KirBac1.1 (Kuo et al., 2003). Since the rod-like densities forming the central ion-conduction pathway are not well defined at the lumenal and cytoplasmic vestibules of the channel pore, it is conceivable that the pore-lining helices might be longer than predicted by hydrophobicity analysis and thus, explains their appearance in the present structure. Finally, the predicted α-helical TM5 segment (residues 2441–2463) would correspond to the partially resolved outer helix in our structure of IP3R1 (Figure 5 and Figure S5B, C).
The Lumenal Turrets
On the lumenal face of the channel we observe structures resembling the outer pore turrets identified in Kir channels (Tao et al., 2009) (Figure 5B, C, Movie S1). We propose that these turret densities are formed by the extended loop between the TM5 and TM6 segments in IP3R sequence (residues 2464–2529), which is absent in RyRs (Figure S5B, C). While the precise function of the turrets in IP3Rs remains to be elucidated, it is notable that the turret sequence in IP3R1 contains many Asp and Glu residues. The high density of negative charges is thought to enable Ca2+ release channels to regulate local concentration of permeant ions at the pore entryway. The turret region also includes two glycosylation sites (Asn2476 and Asn2504) (Figure S5B)(Michikawa et al., 1994). An N-glycosylation site located in the outer turret between the S5 and pore helices has been shown to play a critical role in trafficking of HCN channels to cell surface membrane (Much et al., 2003).
The Cytoplasmic Region
The large CY structural variations shown in the 3D variance map (Figure 4B) combined with the marginal resolution raise too many uncertainties to make reliable α-helix predictions in this region. In addition, available crystal structures for the N-terminal domains (Bosanac et al., 2002; Bosanac et al., 2005) do not unambiguously dock into our structure, due to their small size and the lack of secondary structure elements or other supporting structural information to anchor the docking. For this reason, there are few details we can use to better characterize the CY region, other than to make the observation that it has a notably different architecture than the RyR1 CY region (Ludtke et al., 2005; Samso et al., 2005; Serysheva et al., 2008).
The column region represents the only direct link between structural elements of the CY and the pore-forming domains in our cryo-EM structure of IP3R1 (Movie S1). High structural variability found to be associated with the bridging densities of the column region (Figure 4B and Movie S2) suggests that this region is quite mobile in the present structure. Although the protein sequence comprising the column densities remains to be identified, the ~160-residue C-terminus is a potential candidate along with the N-terminal sequence preceding the TM region. While the limited resolution in the current study did not allow identification of secondary structure elements in the column region, based on the secondary structure prediction, the C-terminus of IP3R1 represents a helical structure. It interacts with several modulatory proteins that enhances IP3-binding sensitivity of the N-terminus and affects the channel gating (Foskett et al., 2007; Mikoshiba, 2007). This implies an allosteric role of the C-terminus in transmitting the signal from the IP3-binding region to the channel pore. Thus, the structure comprising this sequence is expected to be mobile to be able to adapt to different ligand-induced conformations.
Conclusions
In this work we have unambiguously defined the 3D architecture of the IP3R1 channel by obtaining higher contrast cryo-EM images than were previously possible. Resolution is sufficient to provide initial insights into the structure of the ion-conduction pathway, and relate it to other ion channels. The TM architecture of IP3R1 captured in a closed ligand-free conformation is similar to that seen in closed K+ channels (Doyle et al., 1998; Kuo et al., 2003; Tao et al., 2009) and, combined with previously determined structures of the Ca2+ release channel (Ludtke et al., 2005; Samso et al., 2005), may serve as a prototype for this class of membrane proteins. The structure of the CY region encompassing mostly the N-terminal portion of the IP3R1 molecule reflects low structural homology with other ion channels and provides plausible explanations for crucial differences in their function. The present structure suggests that the gating of IP3R1 likely shares a fundamentally similar mechanism with other ion channels, including RyR1 Ca2+ release channel and K+ channels. This involves movements of cytoplasmic ligand-binding domains that are transmitted via the flexible stalk densities of the column region to the channel pore. Further details of the function of IP3R1 will require studies at higher resolution and in different functional states. The improved image contrast obtained in the current studies indicates that this goal is now within reach.
MATERIALS and METHODS
Protein preparation
IP3R1 was purified by immunoaffinity chromatography as described previously (Serysheva et al., 2003), with the following modifications. Microsomal membranes (2–3 mg/ml) were prepared from rat cerebellum (Mignery et al., 1990) and solubilized at 4º C, using 2% (w/v) CHAPS in 50 mM Tris-HCl (pH 7.4), containing 150 mM NaCl, 1 mM DTT, 1 mM EDTA and protease inhibitors (Buffer A). Insoluble membranes were removed by centrifugation at 100,000×g for 1 hour using the Ti45 rotor (Beckman Coulter). The solubilized IP3R1 channels were mixed with CNBr-activated Sepharose 4B beads coupled with the T433 antibody (Kaznacheyeva et al., 1998), incubated at 4º C, and eluted with buffer A containing 0.4% CHAPS and 0.2 mg/ml of the immunogenic peptide corresponding to the 19 C-terminal residues of the rat IP3R1. The purified protein was concentrated in buffer D (buffer A + 0.4% CHAPS) to a final concentration of ~1 mg/ml using an Amicon Ultracel-100 centrifugal filter unit (Millipore). Purified IP3R1 was analyzed by SDS-PAGE and on a Western blot using specific antibody against IP3R1. Immunoblotting of the purified IP3R1 with antibodies raised against IP3R2 and IP3R3 did not show cross-reaction, or such contamination was below the detection limit. PAGE gel and western blot imaging were performed using the Odyssey Infrared System (Li-COR Bioscience).
Functional characterization
To test the functionality of the purified protein, Ca2+-flux measurements were performed with the purified IP3R1 channels reconstituted into lipid vesicles (liposomes) using a previously described procedure (Moiseenkova-Bell et al., 2008). Membrane-impermeant Ca2+-sensitive fluorescent dye, Fura-2 (Invitrogen), was used in time-based fluorescence measurements to monitor IP3-induced Ca2+ efflux from Ca2+-loaded lipid vesicles. In brief, egg phosphatidylcholine (PC; Sigma-Aldrich) was solubilized with 400 mM OG in reconstitution buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM Ca2+. Purified IP3R1 channels (~1 mg/ml) were mixed with the solubilized lipid and dialyzed against reconstitution buffer for 2–4 days at 4º C. To remove Ca2+ outside the vesicles, the Ca2+-loaded vesicles were passed through Econo-Pac 10DG columns (BioRad) and treated with a chelating resin (Chelex-100, Bio-Rad). Fura-2 and channel-specific ligands were prepared in metal-free form using Chelex-100. The resulting vesicles were transferred into 4-ml cuvette, and 5 μM Fura-2 was added to start the Ca2+ flux experiment. Fluorescence measurements were performed with a spectrofluorometer SLM8000 (Olis). Fluorescence emission at 510 nm was recorded continuously while agonist was added step-wise to the cuvette. Ionomycin (3 μM) was used to determine maximum Ca2+ efflux from liposomes.
Cryo-specimen preparation and cryo-EM imaging
In contrast to ryanodine receptor (RyR), a related channel whose higher order structure remained stable in the presence of detergent (Ludtke et al., 2005; Samso et al., 2005), tetrameric IP3R1 channels isolated in a similar manner are significantly less stable and tend to dissociate and aggregate producing heterogeneous particle populations. Therefore, throughout our structural studies it was necessary to vitrify the detergent-extracted protein immediately upon purification to minimize protein degradation. A long delay or a freeze-thaw cycle can easily lead to degraded protein. In addition, 5% sucrose was added to preserve the structural integrity of channel particles. The choice of detergent is also critical to obtaining optimal cryo-EM data. Imaging must be performed as far below CMC as possible to avoid the appearance of micelles in the cryo-EM images, also impacting the choice of detergent. The purified channels (3 μl of 0.5 mg/ml protein) in buffer D containing 5% sucrose and 1 mM EGTA were plunge frozen on Quantifoil holey grids (Quantifoil Micro Tools GmbH) covered with a freshly prepared thin continuous carbon film. Grids were glow-discharged for 5 seconds immediately before the application of the protein. Vitrification was performed at 80–100% humidity, 20°C using a semi-automated vitrification device, Vitrobot (FEI). Using these conditions of vitrification as the starting point, further cryospecimen optimization was performed by conducting multiple vitrification trials; evaluating the results with electron cryo-microscope and using obtained information to improve successive trials of cryo-specimen preparation. Thus, final conditions for vitrification were varied to some degree in each prep while still remaining close to the starting conditions. The critical aspect was to continue the screening process until contrast had been optimized. Given the very limited lifetime of the purified specimen, this process was completed as efficiently as possible. Images of ice-embedded channels were taken at 60,000× nominal magnification on a JEM2010F electron cryomicroscope operated at 200 keV with a Gatan liquid nitrogen cryoholder and a Gatan 4k × 4k CCD camera. The specimen dose was ~18 electrons/Å2 per CCD frame. Images were acquired at the defocus range of 1.4–3.5 μm underfocus.
Image-processing and 3D reconstruction
Particles were selected from individual CCD frames, and contrast transfer function parameters were determined using standard methods in EMAN (Ludtke et al., 2004; Ludtke et al., 2005). Unlike our previous studies on a continuous carbon substrate, the ice-embedded IP3R1 particles have a fairly uniform distribution of orientations, providing isotropic resolution (Figure S2C). Iterative reconstruction consists of reference-based classification of particles, class averaging with contrast transfer function correction and 3D model construction. Refinement in EMAN1 followed the same standard procedures described previously (Ludtke et al., 2004; Ludtke et al., 2005). Iterative reconstruction with 37,231 particles from 869 CCD frames was continued until convergence was achieved. To evaluate model bias, the structure was refined from multiple starting models and the results compared. The map resolution was assessed based on the 0.5 Fourier shell correlation (FSC) criterion and filtered to optimize resolvability without inducing noise (Figure S2D).
Given the controversial history of the IP3R1 structure, as an additional validation, we processed the same particle set from scratch, including CTF determination, initial model generation, refinement and variance map production, using EMAN2 (Tang et al., 2007), which has a new initial model generation method and a new CTF correction strategy which can compensate for the presence of continuous carbon substrates. The results from the two completely independent reconstructions agree to within the limits of resolution and model variance. Both reconstructions using the current image set agree completely about the quaternary structure of the channel, which is not the case when we attempt a reconstruction using the images from our previously published structure (Serysheva et al., 2003). It now seems very likely that model bias was prominent in our earlier study due to high noise levels (Figure S2A). The present study passes much more stringent tests for self-consistency in the reconstruction. This overall shape also agrees well with recent low-resolution results in negative stain (Hamada et al., 2003). While the resolution remains marginal for secondary structural interpretation, the interpretability of the TM helix arrangements in terms of other channel structures lends credence to our interpretation even at this level of detail. The structure used for interpretation and presented in the figures in the body of the paper is the structure solved using the proven methods from EMAN1.
3D variance map
The EMAN1 variance map was produced using calculateMapVariance.py (Chen et al., 2008), which makes use of a variant of the bootstrap method suggested by Penczek (Penczek et al., 2006). In brief, 100 subsampled 3D models were produced from randomly selected particle images, and the 3D average and 3D standard deviation maps were computed from them. The 3D standard deviation map was further filtered to ~15 Å. 3D variance analysis in the presence of symmetry is subject to exaggeration within a few pixels of the symmetry axis, but our observed high variability regions are sufficiently far from the axis that this is not a factor.
Segmentation and interpretation of cryo-EM density map
We applied the same connectivity-based, symmetry preserving segmentation algorithm applied to RyR1 (Ludtke et al., 2005) to identify subunit boundaries in IP3R1 structure. This algorithm segments the structure into a number of high-density regions, and then determines the strength of the connectivity between these regions, establishing segmentation where the weakest connectivities represent the boundaries between subunits. In the TM domain, this separation is unambiguous, however in the CY domain, variation of the segmentation parameters can produce a few possible segmentations. We selected the segmentation, which is consistent with segmentation of RyR1 (Serysheva et al., 2008), but at this resolution, this result is strictly hypothetical.
The 3D density map was visualized using the UCSF Chimera package (Goddard et al., 2005) and AMIRA (Mercury Computer System). At ~1 nm resolution it is possible to identify well defined α-helices, but this must be done with some caution below 8 Å resolution (Baker et al., 2007; Baker et al., 2010). Secondary structure elements (SSE) were identified by visual interpretation of cryo-EM densities and using quantitative estimation with SSEhunter, which is a part of the AIRS package (Baker et al., 2007; Baker et al., 2010). At 6–8 Å resolutions, SSE, i.e. α-helices and β-sheets appear as rodlike densities and thin planes, respectively. At resolutions of 8–10 Å, results are far more ambiguous, and β-sheet identification cannot be performed with confidence. In visually examining the TM region, 6 rod-like densities can be observed. This is confirmed by SSEhunter, which also identifies these regions as α-helices. Similar to our earlier RyR structure (Ludtke et al., 2005; Serysheva et al., 2008), identified helices are only those where both methods agreed. Errors in rotational alignment of the particles will have a decreasing impact on structural features near the center of mass, and along the Z-axis. This is likely possible for relative clarity of the pore-lining helices in both this channel and our earlier structure of RyR1 channel (Ludtke et al., 2005; Serysheva et al., 2008). Initial fitting of the X-ray structures of K+ channels into the IP3R1 TM region was carried out manually and then refined with Chimera.
Sequence alignment and secondary structure prediction
Multiple sequence alignments were performed using ClustalW2 (Larkin et al., 2007). Hydrophobicity analysis and secondary structure prediction were carried out with several packages available through PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) and the Biology Workbench (http://workbench.sdsc.edu).
Supplementary Material
Highlights.
3D cryo-EM structure of IP3R in the closed state at subnanometer resolution
Molecular architecture of ion-channel conduction pore
Discovery of turret densities on the channel’s lumenal surface
Structural flexibility of the CY region in the absence of IP3 and Ca2+
Acknowledgments
We thank G. A. Mignery, D. J. Bare and J. T. Maxwell for providing the IP3R1 protein for initial studies, D. F. Boehning for help with production of IP3R1-specific antibody, T. G. Wensel for help with the Ca2+ flux experiments; J. Zhang for assistance with the 3D statistical variance analysis; M. L. Baker for help with the SSEhunter program; R. Rochat for assistance with graphics; movie S1 was produced by Matthew Dougherty. This research is supported by grants from NIH (P41RR002250, R01GM072804, R01GM080139 and RGM072804Z) and MDA Research Grant 88677.
Footnotes
Supplemental information includes five figures, table and movies and can be founded with this article online.
Author contributions
T.P.T, Q.T.N and I.I.S performed the protein purification; T.P.T and V.Yu.M.-B. performed the Ca2+ flux experiments; Q.T.N. assisted with a cryo-specimen preparation; I.I.S. took single-particle cryo-EM images; I.I.S. and S.J.L. performed the single-particle reconstruction; I.I.S., S.J.L. and W. C. interpreted the map; S.J.L., W.C. and I.I.S. wrote the paper with contributions from all coauthors.
Data deposition
Cryo-EM density map of IP3R1 has been deposited to the EMDB (http://www.emdatabank.org) with accession code EMD-5278.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Zhang J, Ludtke SJ, Chiu W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nat Protoc. 2010;5:1697–1708. doi: 10.1038/nprot.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehning D, Joseph SK. Direct association of ligand-binding and pore domains in homo- and heterotetrameric inositol 1,4,5-trisphosphate receptors. Embo J. 2000;19:5450–5459. doi: 10.1093/emboj/19.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosanac I, Alattia JR, Mal TK, Chan J, Talarico S, Tong FK, Tong KI, Yoshikawa F, Furuichi T, Iwai M, et al. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Yamazaki H, Matsu-Ura T, Michikawa T, Mikoshiba K, Ikura M. Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor. Mol Cell. 2005;17:193–203. doi: 10.1016/j.molcel.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Chan J, Whitten AE, Jeffries CM, Bosanac I, Mal TK, Ito J, Porumb H, Michikawa T, Mikoshiba K, Trewhella J, Ikura M. Ligand-induced conformational changes via flexible linkers in the amino-terminal region of the inositol 1,4,5-trisphosphate receptor. J Mol Biol. 2007;373:1269–1280. doi: 10.1016/j.jmb.2007.08.057. [DOI] [PubMed] [Google Scholar]
- Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- Chen DH, Luke K, Zhang J, Chiu W, Wittung-Stafshede P. Location and flexibility of the unique C-terminal tail of Aquifex aeolicus co-chaperonin protein 10 as derived by cryo-electron microscopy and biophysical techniques. J Mol Biol. 2008;381:707–717. doi: 10.1016/j.jmb.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Morris SA, Nerou EP, Taylor CW, Morris EP. Domain organization of the type 1 inositol 1,4,5-trisphosphate receptor as revealed by single-particle analysis. Proc Natl Acad Sci U S A. 2003;100:3936–3941. doi: 10.1073/pnas.0536251100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T, Yoshikawa S, Miyawaki A, Wada K, Maeda N, Mikoshiba K. Primary structure and functional expression of the inositol 1,4,5- trisphosphate-binding protein P400. Nature. 1989;342:32–38. doi: 10.1038/342032a0. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Hamada K, Terauchi A, Mikoshiba K. Three-dimensional rearrangements within inositol 1, 4, 5-trisphosphate receptor by calcium. J Biol Chem. 2003;278:52881–52889. doi: 10.1074/jbc.M309743200. [DOI] [PubMed] [Google Scholar]
- Hirata M, Narumoto N, Watanabe Y, Kanematsu T, Koga T, Ozaki S. DL-myo-inositol 1,2,4,5-tetrakisphosphate, a potent analog of D-myo-inositol 1,4,5-trisphosphate. Mol Pharmacol. 1994;45:271–276. [PubMed] [Google Scholar]
- Jiang QX, Thrower EC, Chester DW, Ehrlich BE, Sigworth FJ. Three-dimensional structure of the type 1 inositol 1,4,5-trisphosphate receptor at 24 Å resolution. Embo J. 2002a;21:3575–3581. doi: 10.1093/emboj/cdf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002b;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Kaznacheyeva E, Lupu VD, Bezprozvanny I. Single-channel properties of inositol 1,4,5-triphosphate receptor heterologously expressed in HEK-293 cells. J Gen Physiol. 1998;111:847–856. doi: 10.1085/jgp.111.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 Å Resolution by Single Particle Electron Cryomicroscopy. Structure (Camb) 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Serysheva II, Hamilton SL, Chiu W. The pore structure of the closed RyR1 channel. Structure (Camb) 2005;13:1203–1211. doi: 10.1016/j.str.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant JS, Taylor CW. Rapid activation and partial inactivation of inositol trisphosphate receptors by inositol trisphosphate. Biochemistry. 1998;37:11524–11533. doi: 10.1021/bi980808k. [DOI] [PubMed] [Google Scholar]
- Michikawa T, Hamanaka H, Otsu H, Yamamoto A, Miyawaki A, Furuichi T, Tashiro Y, Mikoshiba K. Transmembrane toplogy and sites of glycosylation of the inositol triphosphate receptor. J Biol Chem. 1994;269:9184–9189. [PubMed] [Google Scholar]
- Mignery GA, Newton CL, Archer BT, Sudhof TC. Structure and expression of the rat inositol 1,4,5-trisphosphate receptor. J Biol Chem. 1990;265:12679–12685. [PubMed] [Google Scholar]
- Mignery GA, Sudhof TC, Takei K, De Camilli P. Putative receptor for inositol 1,4,5-trisphosphate similar to ryanodine receptor. Nature. 1989;342:192–195. doi: 10.1038/342192a0. [DOI] [PubMed] [Google Scholar]
- Mikoshiba K. IP3 receptor/Ca2+ channel: from discovery to new signaling concepts. J Neurochem. 2007;102:1426–1446. doi: 10.1111/j.1471-4159.2007.04825.x. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Much B, Wahl-Schott C, Zong X, Schneider A, Baumann L, Moosmang S, Ludwig A, Biel M. Role of subunit heteromerization and N-linked glycosylation in the formation of functional hyperpolarization-activated cyclic nucleotide-gated channels. J Biol Chem. 2003;278:43781–43786. doi: 10.1074/jbc.M306958200. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Yang C, Frank J, Spahn CM. Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Samso M, Wagenknecht T, Allen PD. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat Struct Mol Biol. 2005;12:539–544. doi: 10.1038/nsmb938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Hamada K, Ogura T, Miyazawa A, Iwasaki K, Hiroaki Y, Tani K, Terauchi A, Fujiyoshi Y, Mikoshiba K. Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. J Mol Biol. 2004;336:155–164. doi: 10.1016/j.jmb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Schug ZT, Joseph SK. The role of the S4-S5 linker and C-terminal tail in inositol 1,4,5-trisphosphate receptor function. J Biol Chem. 2006;281:24431–24440. doi: 10.1074/jbc.M604190200. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Bare DJ, Ludtke SJ, Kettlun CS, Chiu W, Mignery GA. Structure of the type 1 inositol 1,4,5-trisphosphate receptor revealed by electron cryomicroscopy. J Biol Chem. 2003;278:21319–21322. doi: 10.1074/jbc.C300148200. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc Natl Acad Sci U S A. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 Å resolution. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K, Miyauchi H, Furuichi T, Michikawa T, Mikoshiba K. Critical regions for activation gating of the inositol 1,4,5-trisphosphate receptor. J Biol Chem. 2003;278:16551–16560. doi: 10.1074/jbc.M300646200. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.