Figure 1. Increasing modeling difficulty of targets in GPCR Dock 2010 correlated with the decrease in prediction accuracy.
(A) Sequence and structural similarity of the assessment targets to the available homology modeling templates: crystal structures of bovine rhodopsin (Palczewski et al., 2000), opsin (Park et al., 2008; Scheerer et al., 2008), human β2AR (Cherezov et al., 2007), turkey β1AR (Warne et al., 2008), and human A2AAR (Jaakola et al., 2008). For CXCR4, the highest sequence similarity template (β1AR) is only ~25% identical and structurally quite dissimilar from the target. For D3, β1AR and β2AR represent relatively high sequence and structural similarity templates. The values were obtained by comparison with PDB entries 1u19 (bRho), 2vt4 (β1AR), 2rh1 (β2AR), 3eml (A2AAR) and 3cap (opsin).
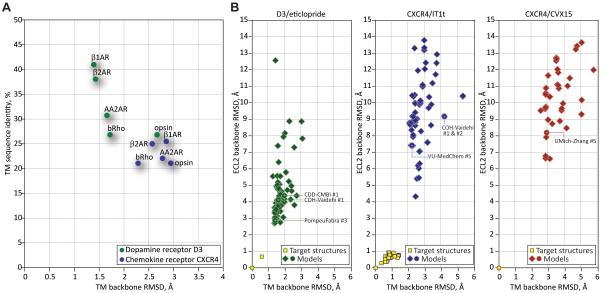
(B) Scatter plots of model TM domain RMSD and ECL2 RMSD values comparison with the target structures for the three assessment targets. Source data for these plots can be found in Supp. Table S1.