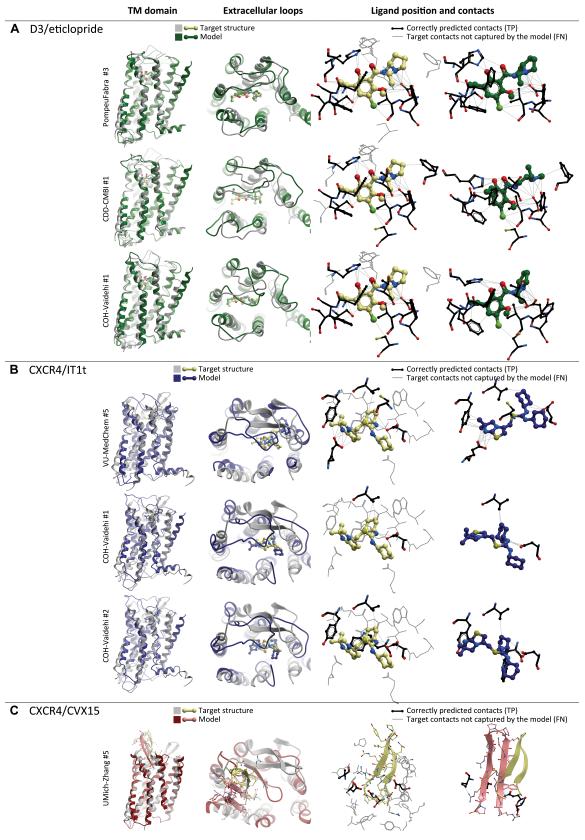
Figure 8. Top scoring models of the three GPCR Dock 2010 assessment targets.
These and other models can also be viewed in interactive mode on the assessment results page at http://ablab.ucsd.edu/GPCRDock2010/.
(A) Most accurate models of D3/eticlopride complex: up to 60% of ligand-pocket contacts are reproduced correctly, the ligand pose and orientation closely resemble those observed in the X-ray structure.
(B) Top scoring models of the CXCR4/IT1t complex. Although the models reproduce the critical hydrogen bonding interaction of the ligand with Asp97, and in one case even with Glu288, very few other contacts are captured, and the ligand orientation is only approximately correct.
(C) Top-scoring model of the CXCR4/CVX15 complex. Relative orientation of the peptide and its β-hairpin fold are reproduced with some degree of similarity to the experimental structure. Only a few residue contacts are captured.