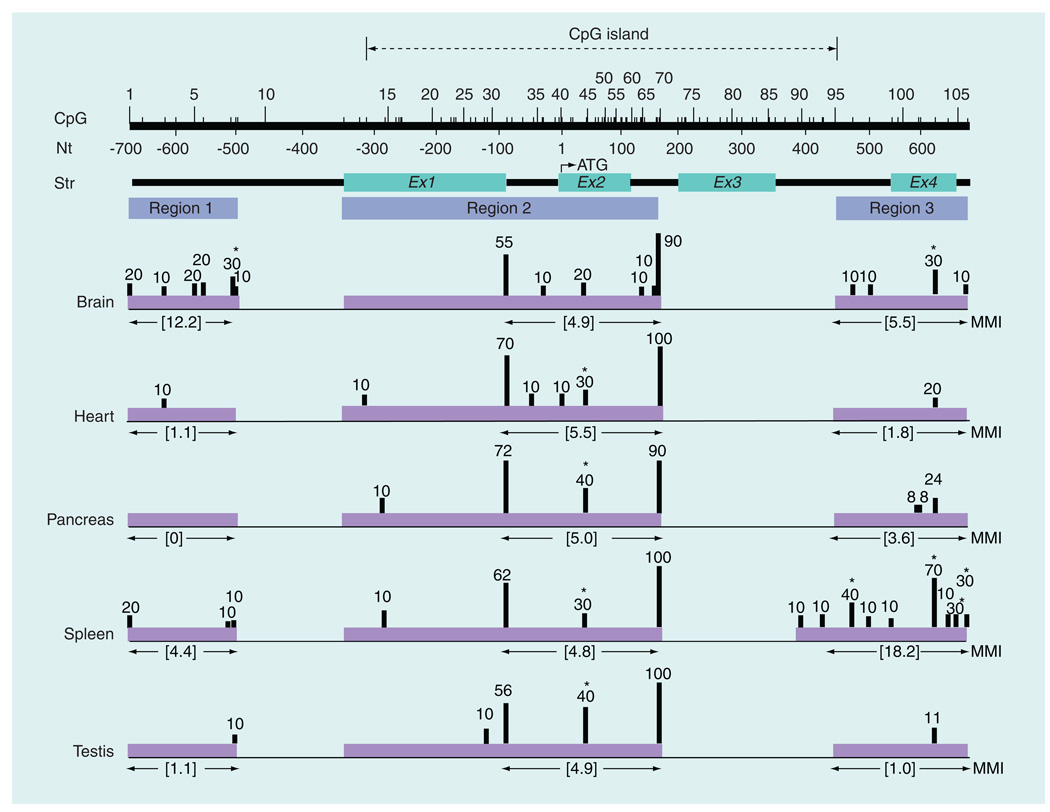
Figure 1. CpG methylation profile of rat Isyna1 in various tissues.
CpG methylation profile was determined in five (male) rat tissues – brain cortex, heart, pancreas, spleen and testis. The CpG island (dotted lines with arrowheads), number and position of each CpG residue, nucleotide position and gene structure are indicated on the top of the figure. The CpG island, determined using European Molecular Biology Open Software Suite (EMBOSS) CpGPlot [102], extends from CpG13–CpG95. Regions 1, 2 and 3 represent amplicons used for CpG analysis; region 2 is composed of three overlapping amplicons. Analyzed regions are shown in purple shaded horizontal blocks with the MMI values directly below each block. Methylation levels (%) are denoted by the height of the vertical bars and are based on the percentage of clones analyzed. For CpG31, the methylation level was averaged from two overlapping amplicons. CpG sites that are significantly differentially methylated in tissues are marked by an asterisk. Note, in particular, the pronounced differences in methylation patterns in the brain cortex in region 1 and in spleen in region 3. Except for CpG10 and CpG71–94, the status of all CpG residues spanning a 1.35-kb region from nucleotides −700 bp of the promoter to +650 bp in intron 4 was examined.
MMI: Mean methylation index; Nt: Nucleotide; Str: Structure.