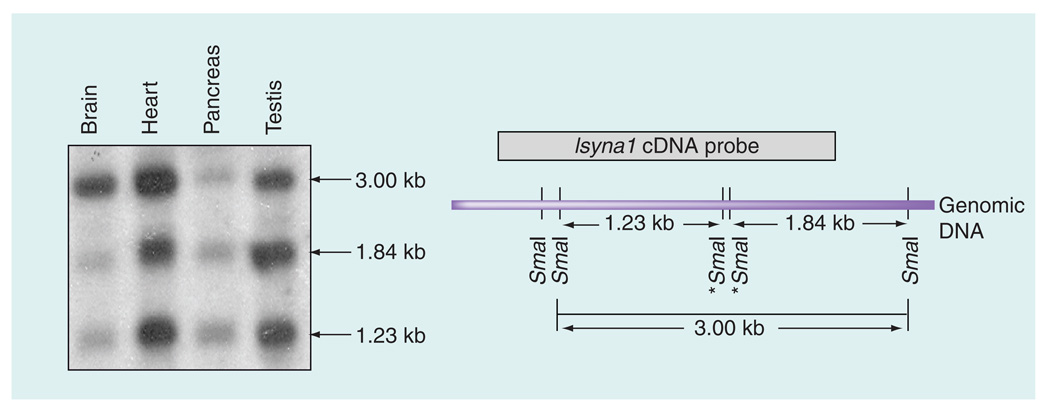
Figure 3. Southern blot analysis of SmaI-digested Isyna1 genomic DNA in different rat tissues.
A total of 20 µg of genomic DNA from four rat tissues (brain cortex, heart, pancreas and testis) was first digested with EcoRI and HindIII, followed by SmaI, resolved on 1% agarose gels, Southern blotted onto a Zeta-Probe GT membrane (BioRad, CA, USA) and probed with a P32-labeled Isyna1 cDNA. The SmaI restriction map of the rat Isyna1 genome is shown on the right. The 1.23-kb and 1.84-kb bands represent completely digested SmaI fragments while the 3.0-kb fragment is due to partial digestion resulting when one or both internal SmaI sites (marked by asterisks) are methylated. The probe detects 1.23-, 1.84- and the 3.00-kb fragments. Differential methylation was inferred by comparing the intensity of the 3.00 kb fragment with that of the other two restriction digest fragments on the blot. Note the striking difference in band intensities between the brain cortex (more methylated) and testis (less methylated).