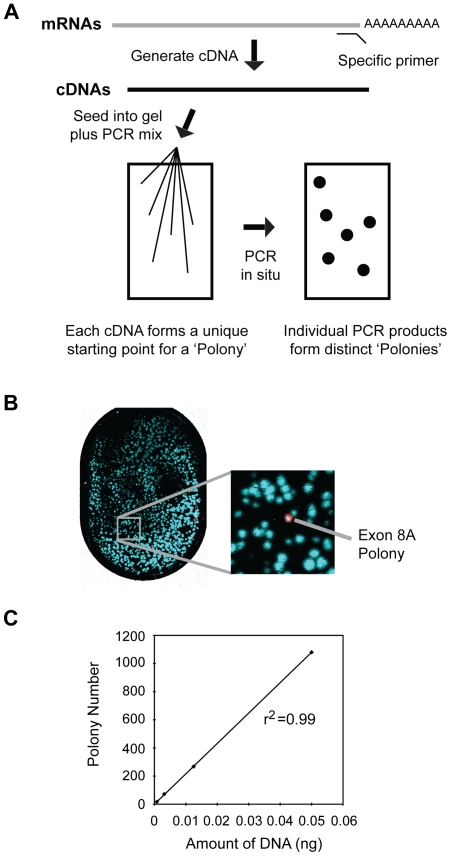
Figure 2. Sensitivity and linearity of the polony assay.
(A) Outline of polony assay. Gene-specific primers are used to convert mRNA to cDNA, which is then seeded into a thin acrylamide gel containing primers, enzymes and other components necessary for in situ PCR, which results in the formation of “PCR colonies”, or polonies. The polonies are fixed in the gel, but can be interrogated by sequential hybridization with specific fluorescently labeled oligonucleotides. (B) Sensitivity of polony assay. Plasmids containing cDNAs for wild type c-myb or the 8A splice variant were mixed in a ratio of 5,000∶1. After in situ PCR, the slide was hybridized sequentially with fluorescently labeled probes specific for exon 8, exon 8A or exon 11. The full polony slide is shown at left and the enlargement at right shows a single detected exon 8A-containing splice variant (white). In the false color image the polonies containing only exons 8 and 11 appear Cyan in color, the polony containing all three exons appears white. (C) Linearity of polony assay. Different amounts of wild-type c-myb plasmid were used as template in the polony assay and the number of polonies detected with the exon 11 probe was plotted against amount of plasmid template used in the assay. The assay was very linear, as shown by the correlation coefficient r2, which was 0.99.