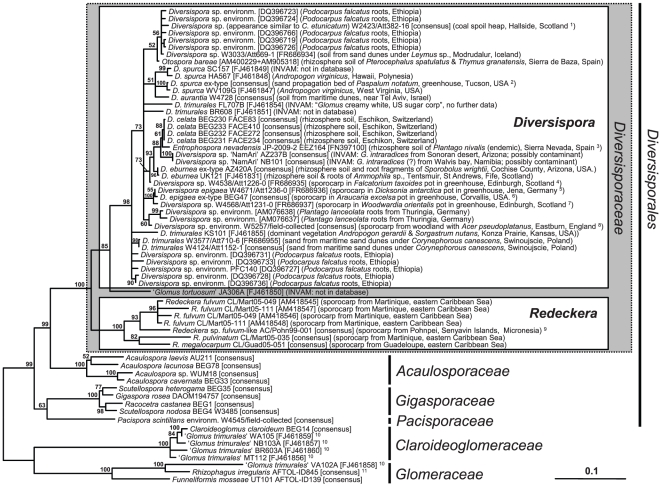
Figure 2. Phylogenetic tree of Diversisporaceae computed from the extended dataset, including environmental nuclear rDNA sequences.
RAxML maximum likelihood analysis with bootstrap support shown at the branches; topologies with support below 50% were collapsed to polytomies. The tree is rooted with representatives of the Glomerales. The scale bar indicates proportional substitutions per site. Except for very short environmental SSU rDNA sequences that distorted the tree topology, all Diversisporaceae sequences which were available from the public databases were used and have the following origins: 1 the specimen from which this sequence was derived has Claroideoglomus etunicatum-like spore morphology; soil from a re-vegetated coal spoil heap, beneath Salix sp. and associated weeds, which included Plantago major, P. lanceolata, Fragaria vesca and various grasses; 2 Fazio's Greenhouse, from M. Pfeiffer's pot culture no. 157, Building 42-2R, University of Arizona; 3 other plants reported at the soil sampling location were Alchemilla fontqueri and Senecio elodes (both endemic) and Sorbus hybrid (non-endemic); 4 fungus with an appearance similar to a ‘large-spored D. epigaea’, from a temperate greenhouse of Royal Botanical Garden Edinburgh, Plant No. 842581 H; 5 immature spores; from fern house of Botanical Garden Jena (the plant was transferred to Jena from the botanical garden of the Wilhelma, Stuttgart, Germany); 7 Diversispora epigaea-like spores; temperate greenhouse of Royal Botanical Garden Edinburgh, the pot also contained an Oxalis sp. as a weed; 6 tropical greenhouse at the USDA-ARS horticultural research station; 8 sporocarp from litter layer of semi natural woodland, with associated understory, including an Allium sp.; 9 this sequence most likely represents a species distinct from Redeckera fulvum, therefore it is annotated here as ‘R. fulvum-like’; 10 sequences annotated as ‘D. trimurales’, from the same submission as the three sequences (FJ461851,54,55) that cluster in Diversispora, but clearly falling in distinct families; 11 culture published as GINCO4695rac-11G2 from the AFTOL project, but lacking further information.