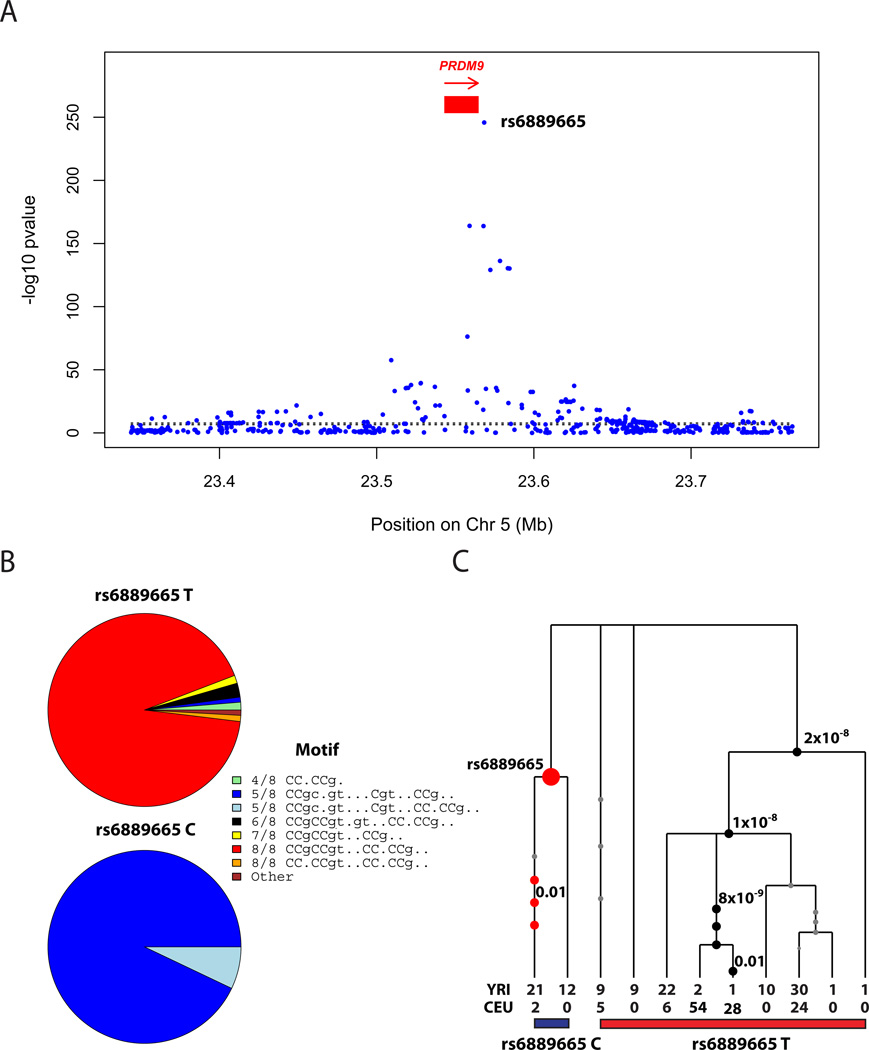
Figure 2.
Association of PRDM9 genetic variation with hotspot activity. (A) A GWAS measuring association of the “African-enrichment” (AE) phenotype shows a single genome-wide significant peak at PRDM9, with rs6889665 the best associated SNP. (B) Relationship between alleles at the rs6889665 and predicted binding target of the PRDM9 zinc finger array9 for West African and European samples. The alleles are grouped into 8 clusters according to their best-matching region to the 13-bp motif, and annotated by the number of bases matching the motif. The African-enriched rs6889665 “C” allele always co-occurs with motifs with a poor (5/8) match to the 13-mer. (C) Gene tree32 of the LD block containing the PRDM9 ZF array (Methods); numbered circles show SNPs and significant P-values for association, after conditioning on rs6889665.