Figure 2.
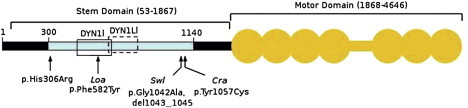
Schematic Representation of Human DYNC1H1
The N-terminal region of DYNC1H1 is represented by a horizontal black bar, and the stem domain (amino acids 53–1867) is indicated by a bracket above. Residues involved in DYNC1H1 dimerization (300–1140) are shown by a light blue bar; open boxes represent binding regions for intermediate (DYN1I; residues 448–703, solid line) and light-intermediate (DYN1LI; residues 651–802, broken line) chains, respectively. The motor domain (amino acids 1868–4646) is indicated by the orange shaded area; the seven ATPase domains are represented by circles, whereas the horizontal bar indicates the stalk region. The equivalent positions of mutations in three mouse models, Loa, Swl, and Cra1, are shown below the figure along with the p.His306Arg mutation identified in the family reported here. Note that the human protein contains two additional glycine residues at position 7 relative to mouse Dync1h1, that is numbering of equivalent residues in human DYNC1H1 is 2 higher than in mouse models. Phe582 is within a highly conserved domain responsible for binding the dynein intermediate chains as well as homodimerization.4 The 9 bp Swl deletion and Cra p.Tyr1057Cys mutation are outside the dynein intermediate-chain binding region but within the putative homodimerization domain.4 Numbering of amino acids is taken from Tynan et al.16 and the UniProtKB entry for human DYNC1H1 (identifier Q14204).
