Abstract
The search for improved molecular cancer diagnostics is a challenge for which systems approaches show great promise. As is becoming increasingly clear, cancer is a perpetually-evolving, highly multi-factorial disease. With next generation sequencing providing an ever-increasing amount of high-throughput data, the need for analytical tools that can provide meaningful context is critical. Systems approaches have demonstrated an ability to separate meaningful signal from noise that arises from population heterogeneity, heterogeneity within and across tumors, and multiple sources of technical variation when sufficient sample sizes are obtained and standardized measurement technologies are used. The ability to develop clinically useful molecular cancer diagnostics will be predicated on advancements on two major fronts: 1) more comprehensive and accurate measurements of multiple endpoints, and 2) more sophisticated analytical tools that synthesize high-throughput data into meaningful reflections of cellular states. To this end, systems approaches that have integrated transcriptomic data onto biomolecular networks have shown promise in their ability to classify tumor subtypes, predict clinical progression, and inform treatment options. Ultimately, the success of systems approaches will be measured by their ability to develop molecular cancer diagnostics through distilling complex, systems-wide information into simple, salient, actionable information.
INTRODUCTION
Since President Nixon’s State of the Union address, where he first announced the National Cancer Act of 1971 and effectively declared war on cancer, there have been many important, successes in the treatment and prevention of many cancers (von Eschenbach, 2004). These advances are largely predicated on our increased understanding of cancer etiology, which have recently been accelerated through genomics-enabled science and medicine. However, cancer remains one of the most pervasive causes of death worldwide (Edwards et al., 2010; WHO, 2008), and the enormous difficulty of effectively treating cancers remains. The challenges of harnessing the exponentially increasing amounts of high-throughput data must be met to enable the predictive, preventive, personalized, and participatory medicine envisioned for the future (Hood et al., 2004).
While Knudson’s two-hit model of cancer, in which cancer results from a few accumulated DNA mutations (Knudson, 1971), has explained some cancers, the last decade has provided mounting evidence that more sophisticated, multifactorial models are needed to explain the majority of cancers. Multifactorial models posit that cancer is the result of combined effects from multiple low-penetrance mutations in combination with environmental factors (Fletcher & Houlston, 2010; Fodde & Smits, 2002). Multifactorial models have been greatly informed by our increased ability to assess the individual contributions of the approximately 20,000 human genes (Clamp et al., 2007; Schena et al., 1995). Such analyses have revealed highly diverse mutational patterns across patients, ranging from fewer than 1000 to greater than 100,000 point mutations in sequenced cancer genomes (Durbin et al., 2010; Stratton et al., 2009). Although high-throughput measurements continue to be generated at an exponential rate, interpreting such vast amounts of data poses substantial difficulties. The basis for advances in diagnostic cancer medicine will likely require analytical methods that can extract easily interpretable molecular disease-state indicators from amidst immense biological complexity.
A number of clinically used molecular cancer diagnostics have already provided substantial utility in pre-symptomatic screening, confirmatory diagnosis, and prognosis prediction. Improving upon these established diagnostic markers will require a more comprehensive and accurate picture in terms of both measurement and analysis. We will describe several technical and computational efforts made to overcome these challenges with a focus on integration of heterogeneous information and a prospective outlook of these approaches.
The promise and challenge of preventative and early diagnostics
The value of early diagnostic markers is highlighted by the observation that treatments tend to be more effective the earlier they are applied. While methods are improving, many forms of cancer remain difficult to detect and diagnose. Lung cancer is one such case. Conventional clinical diagnosis is still dominated by symptomatic assessments followed by biopsy confirmation. By the time of symptomatic diagnosis, disease has often spread beyond the initial site of malignancy, which significantly undermines the efficacy of traditional treatments. For lung cancer, where the five-year survival rate varies from 49% for local stage disease to 2% for distal metastasis, two thirds of patients are diagnosed at late stages that are associated with poorer outcomes (Leidinger et al., 2010).
Increasingly, molecular-based cancer diagnostics are aiding standard clinical diagnosis (see Table 1) (AACC, 2010). While molecular markers are mostly used for confirming diagnosis, monitoring patient prognosis and assessing disease subtypes after symptoms are present, other applications for molecular markers are progressively being implemented. These functions include pre-symptomatic screening, guiding treatment options, monitoring treatment efficacy and disease progression and identifying disease recurrence after treatment.
Table 1.
Molecular markers used for clinical cancer diagnosis © 2010 by American Association for Clinical Chemistry. Reprinted with permission from “Tumor Markers” on Lab Tests Online (www.labtestsonline.org). These markers aid in pre-symptomatic diagnosis, disease progression and recurrence monitoring, and treatment guiding.
| TUMOR MARKERS | CANCERS | WHAT ELSE? | WHEN/HOW USED | USUAL SAMPLE |
|---|---|---|---|---|
| AFP (Alpha-feto protein) | Liver, germ cell cancer of ovaries or testes | Also elevated during pregnancy | Help diagnose, monitor treatment, and determine recurrence | Blood |
| B2M (Beta-2 microglobulin) | Multiple myeloma and lymphomas | Present in many other conditions, including Crohn’s disease and hepatitis; often used to determine cause of renal failure | Determine prognosis | Blood |
| CA 15-3 (Cancer antigen 15-3) | Breast cancer and others, including lung, ovarian | Also elevated in benign breast conditions; doctor can use CA 15-3 or CA 27.29 (two different assays for same marker) | Stage disease, monitor treatment, and determine recurrence | Blood |
| CA 19-9 (Cancer antigen 19-9) | Pancreatic, sometimes colorectal and bile ducts | Also elevated in pancreatitis and inflammatory bowel disease | Stage disease, monitor treatment, and determine recurrence | Blood |
| CA-125 (Cancer antigen 125) | Ovarian | Also elevated with endometriosis, some other benign diseases and conditions; not recommended as a general screen | Help diagnose, monitor treatment, and determine recurrence | Blood |
| Calcitonin | Thyroid medullary carcinoma | Also elevated in pernicious anemia and thyroiditis | Help diagnose, monitor treatment, and determine recurrence | Blood |
| CEA (Carcino-embryonic antigen) | Colorectal, lung, breast, thyroid, pancreatic, liver, cervix, and bladder | Elevated in other conditions such as hepatitis, COPD, colitis, pancreatitis, and in cigarette smokers | Monitor treatment and determine recurrence | Blood |
| Chromogranin A (CgA) | Neuroendocrine tumors (carcinoid tumors, neuroblastoma) | May be most sensitive tumor marker for carcinoid tumors | To help diagnose and monitor | Blood |
| Estrogen receptors | Breast | Increased in hormone-dependent cancer | Determine prognosis and guide treatment | Tissue |
| hCG (Human chorionic gonadotropin) | Testicular and trophoblastic disease | Elevated in pregnancy, testicular failure | Help diagnose, monitor treatment, and determine recurrence | Blood, urine |
| Her-2/neu | Breast | Oncogene that is present in multiple copies in 20–30% of invasive breast cancer | Determine prognosis and guide treatment | Tissue |
| Monoclonal immunoglobulins | Multiple myeloma and Waldenstrom’s macroglobulinemia | Overproduction of an immunoglobulin or antibody, usually detected by protein electrophoresis | Help diagnose, monitor treatment, and determine recurrence | Blood, urine |
| Progesterone receptors | Breast | Increased in hormone-dependent cancer | Determine prognosis and guide treatment | Tissue |
| PSA (Prostate specific antigen), total and free | Prostate | Elevated in benign prostatic hyperplasia, prostatitis and with age | Screen for and help diagnose, monitor treatment, and determine recurrence | Blood |
| Thyroglobulin | Thyroid | Used after thyroid is removed to evaluate treatment | Determine recurrence | Blood |
| Other Tumor Markers Less Widely Used | ||||
| BTA (Bladder tumor antigen) | Bladder | Not widely available, but gaining acceptance | Help diagnose and determine recurrence | Urine |
| CA 72-4 (Cancer antigen 72-4) | Ovarian | No evidence that it is better than CA-125 but may be useful when combined with it; still being studied | Help diagnose | Blood |
| Des-gamma-carboxy prothrombin (DCP) | Hepatocellular carcinoma (HCC) | New test; often used along with an imaging study plus AFP and/or AFP-L3% to evaluate if someone with chronic liver disease has developed HCC | To evaluate risk of developing HCC; to evaluate treatment; to monitor for recurrence | Blood |
| EGFR (Her-1) | Solid tumors, such as of the lung (non small cell), head and neck, colon, pancreas, or breast | Not available in every laboratory | Guide treatment and determine prognosis | Tissue |
| NSE (Neuron-specific enolase) | Neuroblastoma, small cell lung cancer | May be better than CEA for following this particular kind of lung cancer | Monitor treatment | Blood |
| NMP22 | Bladder | Not widely used | Help diagnose and determine recurrence | Urine |
| Prostate-specific membrane antigen (PSMA) | Prostate | Not widely used; levels increase normally with age | Help diagnose | Blood |
| Prostatic acid phosphatase (PAP) | Metastatic prostate cancer, myeloma, lung cancer | Not widely used anymore; elevated in prostatitis and other conditions | Help diagnose | Blood |
| S-100 | Metastatic melanoma | Not widely used | Help diagnose | Blood |
| Soluble Mesothelin-Related Peptides(SMRP) | Mesothelioma | Often used in conjunction with imaging tests | To monitor progression or recurrence | Blood |
| TA-90 | Metastatic melanoma | Not widely used, being studied | Help diagnose | Blood |
A prime example of a clinically applied molecular cancer diagnostic is the use of estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2) markers to help determine prognosis and guide treatment in breast cancer patients (Slamon et al., 1987; Slamon et al., 1989). Over two-thirds of all breast tumors are ER positive, and approximately 25–30% are HER2 positive (Slamon et al., 1989). Tumors that are ER positive tend to be HER2 negative, and vice versa (Lal et al., 2005). Typically, ER positive tumors that respond to endocrine therapies tend to be less aggressive, more differentiated, and associated with a more favorable prognosis (Osborne, 1998). HER2 positive tumors tend to be less differentiated, more aggressive and are treated with both chemotherapy and the adjuvant monoclonal antibody Herceptin. The prognostic value of ER, HER2, and, as later discussed, progesterone receptor (PR) expression is great enough that the American Society of Clinical Oncology has recommended that all breast tumors be evaluated for the expression of these proteins (Hicks & Tubbs, 2005; Jones et al., 2010; Lapidus et al., 1998; Mouridsen et al., 2003; Subramaniam & Isaacs, 2005). Such success stories demonstrate that molecular markers in cancer can be highly useful for diagnosis and for guiding therapy.
Despite harnessing decades of research and rapidly increasing high-throughput data, very few newly discovered molecular diagnostic markers demonstrate clinical utility each year. Broadly, advances in clinically applicable molecular cancer diagnostics are predicated on fronts addressing two questions: 1) how well do our measurements represent the physiologically and medically relevant cellular states and 2) how well can we reconstruct predictive models of cellular states based on the data to gain greater understanding and control of the biological mechanisms for improved treatment and reduced side effects?
The Challenge of Representation
A major challenge in the search for clinically robust diagnostic markers is that of attaining adequate measurements. Like a photographer trying to capture a dynamic scene, we seek to construct clear representations of cellular states with only limited measurements. Measurement technologies are faced with two main goals: 1) comprehensive coverage, and 2) accurate representation. While improving consistently, current measurement approaches cannot measure every cellular component accurately and have substantial sources of variability. Clinically applicable measurements are further limited by practical considerations. For example, minimizing patient harm limits the set of biological samples that can be considered. Invasive measurements from tissues are impractical for presymptomatic screening and would ideally be avoided even for post-diagnosis disease monitoring. Bodily fluids are among the few types of samples that represent the dynamic biochemical state of the body and can be collected through non-invasive means. Thus, the majority of currently used molecular markers are molecules found in the blood or urine (Martin et al., 2010). A challenge in biomarker discovery is to find robust indicators of disease that can also accommodate the practical constraints associated with clinical utilization.
ComprehensiveCoverage
Cellular states arise from interactions among myriad functional molecular players. Blood, the most widely used clinical sampling medium, comprises a highly diverse set of proteins and metabolites that span over nine orders of magnitude in concentration (Adkins et al., 2002; van Ravenzwaay et al., 2007), making it an attractive but complex source of health and disease state information. Consequently, comprehensive representation of cellular phenomena necessitates the detection of a broad spectrum of molecules with very large dynamic ranges of sensitivity.
The most mature technology for systematic characterization of cellular states today is in transcriptomics—driven forward by advances in cost and scale of RNA quantification relative to the moderate advances for more difficult challenges such as protein measurement. In total, several hundred thousand transcriptomes have been collected in online public databases such as the Gene Expression Omnibus (Barrett et al., 2009), enabling large-scale associations of thousands of expression measurements with over a hundred disease classes (Huang et al., 2010). Microarrays have been widely used because they offer a global picture of gene transcription (i.e. the transcriptome) in numerous organisms and are relatively easy to use, enabling collection of many samples for individual studies. Despite their utility, microarrays have limitations, including measurement constraints of sensitivity, scope, and dynamic range. Moreover, microarrays are limited in their representation of cellular states as they are almost exclusively used for mRNA gene expression. Emerging direct sequencing methods (e.g. RNA-seq) now promise to greatly advance the information about transcriptomes again (discussed below).
While transcriptomics measurements can provide important information about various biological processes within cells, it is by no means a complete picture. In fact, mRNA abundance alone has been found to account for only 25–30% of the protein abundance variation in a human cancer cell line (Vogel et al., 2010), demonstrating the significance of post-transcriptional regulation and differential protein degradation rates within the cell. Non-coding RNAs, epigenetic modifications and alternative splice variants provide further layers of complexity in the regulation of cellular activities (see Table 2) as well as additional sources for molecular diagnostics. Examples of non-mRNA markers in Table 2 highlight the intricate interplay between genes and other molecules that lead to phenotypic changes in cells. Thus, new experimental approaches that enable standardized, high-throughput, and global measurements of more biological phenomena—such as has been largely achieved with DNA and RNA—are clearly key to advancing the field.
Table 2.
Summary of key molecular modulators of cellular activity in cancers.
| Molecular Modulators | Function in Cancer | Potential Diagnostic Examples | Measurement Approach | References |
|---|---|---|---|---|
| miRNAs |
|
miR-205 correlates with tumor progression in prostate cancer | RNA-seq | (Burchard et al., 2010; Gandellini et al., 2010; Marioni et al., 2008; Mitchell et al., 2008; Ryan et al., 2010; Schaefer et al., 2010; Vasudevan et al., 2007; Wang et al., 2009) |
| lincRNAs |
|
lincRNA HOTAIR contributes to increased malignancy in breast cancer | Custom microarrays, RNA-seq | (Gupta et al., 2010; Huarte et al., 2010; Marioni et al., 2008; Wang et al., 2009) |
| DNA hypermethylations |
|
Hypermethylation of GSTP1 promoter as marker for prostate cancer | MethylC-seq, Reduced representation bisulfate sequencing, MeDIP-seq, MBD-seq, MRE-seq | (Esteller et al., 2001; Harris et al., 2010; Jones & Baylin, 2002; Nakayama et al., 2004; Pomraning et al., 2009) |
| Histone modification |
|
Dimethylation of lysine 4 and acetylation of lysine 18 in histone H3 as marker for prostate cancer recurrence | Mass Spectrometry | (Esteller, 2008; Richon et al., 2000; Vermeulen et al., 2010) |
| SNP |
|
Mutation at P53 codon 72 of the TP53 gene can differentiate response to chemotherapy in head and neck carcinomas | RNA-seq | (Grochola et al., 2010; Marioni et al., 2008; Ryan et al., 2010; Wang et al., 2009) |
| Alternative splice variants |
|
Splice variants of POLB, GPR137, and RUNX2 correlated with breast cancer presence and tumor staging | RNA-seq | (Marioni et al., 2008; Moore et al., 2010; Stickeler et al., 1999; Venables et al., 2008; Wang et al., 2009) |
Measurement technologies typically only capture static snapshots of cellular states, which might lead us to miss diagnostic, time-dependent signatures reflecting dynamic changes within the body. To interrogate the dynamic nature of cellular systems, we need to assemble time series of individual measurements, akin to compiling a flipbook. This challenge can be met through large longitudinal studies wherein measurements are made at regular intervals (Ibrahim et al., 2010), and the patient in effect serves as his or her own control. One example of this phenomenon in cancer is that change in prostate specific antigen (PSA) concentration in a patient’s serum over time can be a better diagnostic for prostate cancer than is the absolute concentration alone (Smith & Catalona, 1994). Another important form of diagnostic test is measuring dynamic responses in molecular markers to an induced perturbation, analogous to measuring blood glucose levels after drinking sugar water in diabetics. Thus comprehensive measurement strategies should explore dynamic changes as well as static signals, using computational methods to help deduce indicative dynamic molecular signatures that reflect altered disease states.
Accurate Representation
In addition to being as comprehensive as possible, measurements must also be correct. Accurate representation of cellular systems facilitates isolation of cohesive signatures that are applicable across the population. Several sources of measurement variability interfere with the ability to gather accurate data. While some sources cannot be avoided, discerning the source of variability is critical to design effectively robust molecular diagnostics.
Noise arises from both the measurement approach (e.g. instrument used, protocol adopted, etc.) and from the heterogeneity across the population, known as technical variation and biological variation, respectively. While biological variation might be addressed with a better understanding of the underlying biological complexities of the diseased state, accurate measurement platforms with appropriate measurement procedures help mitigate technical variation.
Although gene expression microarray technology has demonstrated qualitative consistency (Shi et al., 2006), microarrays are vulnerable to multiple sources of noise. First, the absolute expression values from microarrays are sensitive to probe effects, which vary significantly across platforms (Irizarry et al., 2005). A second vulnerability stems from variation in sample preparation, measurement, and data preprocessing techniques, which contribute to sizable inter-laboratory differences (Irizarry et al., 2005; Shi et al., 2006). The variability across laboratories and microarray platforms stunts the search for robust markers that can be applied in a broad, clinical setting—making community-wide standards critically important.
A more sensitive alternative for measuring transcriptomes is RNA-seq. Compared to microarrays, RNA-seq has been shown to be more reproducible on a single sample basis and more sensitive in detecting differentially expressed genes, allowing the identification of more subtle biological signals (Marioni et al., 2008). Besides offering the potential for increased sensitivity and robustness, direct RNA sequencing allows for a broader dynamic range and provides additional alternative splicing and sequence information, which can be leveraged for improved diagnosis (see Table 2).
A second source of data variability stems from population heterogeneity. Gene expression is a multifactorial function that can be stochastic in nature (Raser & O’Shea, 2005). To this end, the 1000 Genome Project has already yielded important information about population variation within the genome (Durbin et al., 2010). Among their important findings, they reported that each person carries, on average, between 250–300 loss-of-function variants, 50–100 of which have been previously implicated in inherited disorders. The extent of normal variation within each individual presents a significant challenge in identifying the causative mutations in cancer. In addition to the cellular heterogeneity of cancer and other diseases, variation also arises from everyday sources of biochemical fluctuation such as time of day, diet, and even mood (Lesch, 2004; Raser & O’Shea, 2005; Slatter et al., 2006; Winrow et al., 2009). The variation is exacerbated in cancer, wherein genomic instability can cause many ‘passenger’ mutations that do not directly contribute to the disease (Li et al., 2010). Environmental factors can also contribute significantly to disease (de la Chapelle, 2004). Such outcomes suggest that even when we can probe the complete state of a genotype, predicting all of the phenotypic implications might remain beyond reach.
Resolving the Image
The search for diagnostic molecular signatures is obstructed by incomplete coverage and obscured by noise, but piecing together a more complete representation from incomplete snapshots is facilitated by high-throughput measurements. The necessary task in molecular diagnostic discovery is to discern the meaningful biological signals that consistently represent cellular states across the heterogeneous population amidst the technical and biological variation that pervades the measurements.
The plethora of high-throughput data poses both a help and a challenge to developing a meaningful representation of cellular systems. The primary challenge comes from the dangers of multiple hypothesis testing when we typically have vastly more measured variables (e.g. genes) than we do samples. If not carefully guarded against, the large number of potential molecular diagnostics measured can confound genuine signals with correlations that appear by chance, which can mislead rather than enlighten. Increasing sample sizes greatly improves the resolution for this discernment, facilitating the identification of robust signatures for increasingly subtle phenotypic differences. To harness this data, we must turn to machine learning algorithms, which harness the power of statistics and efficient computation to utilize the data by isolating biologically significant associations. Many methods exist for deciphering this information. For example, clustering algorithms have been used to discover molecular subtypes of cancer that are symptomatically indistinguishable but display significantly different expression profiles and respond differently to treatments (Bild et al., 2006; Gatza et al., 2010; Sorlie et al., 2001). Supervised machine learning algorithms such as Support Vector Machine (Ben-Hur et al., 2008; Liu et al., 2005) or Top Scoring Pair approaches (Geman et al., 2004; Price et al., 2007) have isolated sets of molecular identifiers that can distinguish phenotypes with high accuracy.
Ultimately, clinics favor diagnostics that are robust and easily interpreted such that conclusive diagnoses can be made with minimal chance of mistake. Although diagnostic signatures composed of large numbers of genes have demonstrated predictive power (Hedenfalk et al., 2001; van de Vijver et al., 2002), other studies have demonstrated that it is possible to probe cellular systems by monitoring only a few variables (Hwang et al., 2009; Osborne et al., 2005). Practical considerations suggest that diagnostics limited to small numbers of markers that produce quick, unambiguous results will more likely succeed.
The Challenge of Reconstruction
Our understanding of the complexity of biological systems is ever increasing. Despite significant experimental and analytical challenges arising from this complexity, systems approaches have already successfully led to insights into cancer biology and treatment. Important efforts in sequencing the human genome (Durbin et al., 2010; Lander et al., 2001; Venter et al., 2001) and individual cancers (Parsons et al., 2008; Sjoblom et al., 2006) mean that malignant genetic transformations can be studied and modeled in the context of the entire genome. As is becoming increasingly clear through the results of studies including The Cancer Genome Atlas Project (Parsons et al., 2008), cancers arise from multiple low-penetrance mutations, any subset of which lacks a discernable phenotype. Such a distribution of mutations presents enormous challenges for personalized medicine, because it means that simple mutation pattern to treatment correlations are not likely to be effective. Promise lies instead in reconstructing models of the underlying mechanisms based on the representation of the cellular system provided by measurements. Thus, personalized and multi-targeted therapies will ultimately be needed for effective treatment. The increasing emphasis on systems biology approaches that construct and factor in comprehensive networks will be valuable for relating the multiplicity of genetic perturbations to their effects on biomolecular network functions. Systems methods that can predict the effects of modifications are essential to account for diverse molecular causes, where simple correlation methods cannot account for the effects of diverse and rarely repeating mutation combinations. Hence, the creation of more complete and accurate networks associated with human systems is needed to fully realize the potential of network-based biology.
Power of Network-based Context
In modeling the complexity of biological systems, interactions and associations are often organized into networks. Most networks can be categorized into two groups: 1) biochemical networks that are directly mechanistic and typically derived by detailed small-scale experiments and 2) statistical inference networks, which are generally indirectly extrapolated based on mutual information or significance associations of high-throughput data (Edelman et al., 2010; Price & Shmulevich, 2007). Both types of networks can help to inform the reconstruction of cellular models that can aid in the identification of molecular diagnostics. These networks provide a contextual framework in which high-through put a data can be integrated to help discern meaningful signal from noise. In breast cancer, while ER and HER2 status have long-standing clinical utility, several groups have attempted to further classify tumors using expression array data along with network analysis (Culhane & Howlin, 2007; Dati et al., 1990; Perou et al., 2000; Sorlie et al., 2001; Sorlie et al., 2003). In one study, gene expression averaged across protein-protein interaction subnetworks was found to be more accurate and robust in discriminating between breast cancer and normal tissue than gene expression alone (Chuang et al., 2007). In another study, the expression of interacting subnetwork hubs relative to interacting partners improved the prognosis assessment over examination of gene expression without the network context (Taylor et al., 2009).
In addition to improved classification, discriminative subnetworks also demonstrated greater identity overlap across distinct studies, and they have greater enrichment of functionality associated with cancer-related biological processes than do individual gene classifiers. While genes known to be associated with breast cancer (e.g. HER-2/neu, Myc, cyclin D1, etc.) did not display sufficient differential expression across phenotypes to be selected as individual gene classifiers, these cancer genes were included in several discriminative subnetworks as hubs that connected differentially expressed genes (Chuang et al., 2007), suggesting contexts in which known oncogenes can impact the biochemical states of cancer cells.
Biochemical and statistical inference subnetworks also create a context that can better link observed gene expression perturbations with deeper biology. In examining the variability of expression within biochemical networks in samples taken across different phenotypes, increasing network-expression variability has been found in several cancers as they progress to more malignant phenotypes (Eddy et al., 2010). This suggests that certain subnetworks become less tightly regulated as cancers form and progress (Eddy et al., 2010). In one study, subnetwork deregulation could distinguish among normal prostate, primary prostate cancer and metastasizing prostate cancer with greater accuracy than purely expression-based classification algorithms.
Besides aiding classification and augmenting biological insight into cancer, applying biochemical and statistical inference network information can also inform treatment strategies. Understanding network context enables the possibility of treatment through a greater number of mechanisms and provides insight into whether treatment combinations redundantly target network nodes in series or synergistically affect network branches in parallel. Therapeutic agents activating cancer-associated protein-protein interaction subnetworks have demonstrated significant correlation to inhibition of breast cancer cell line proliferation (Bild et al., 2006). Study of metabolic reaction subnetworks has also identified potential therapeutic targets that can disrupt disease processes such as cancer cell metabolism (Duarte et al., 2007; Resendis-Antonio et al., 2010). These studies suggest that targeting cancer-associated subnetworks can ultimately inform rational design of multi-drug treatment regimes, which may be more effective than application of single drugs in isolation (Gatza et al., 2010). The biological context of networks helps to better interpret the numerable changes in gene expression data, aiding us to develop diagnostics with improved outcomes and novel therapeutic strategies.
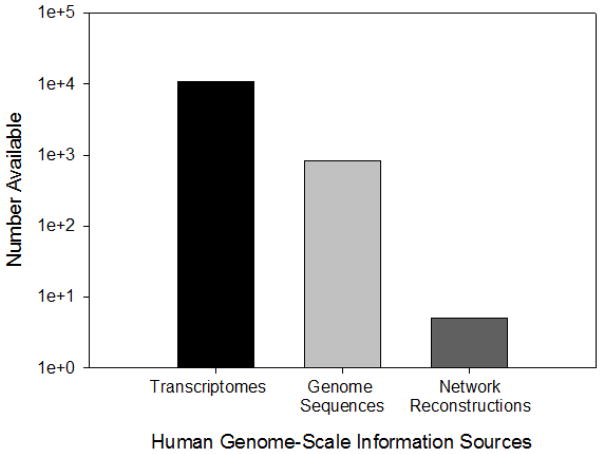
Network Reconstruction
Genome-scale network reconstructions attempt to synthesize a global context of cellular functionality. The greatest obstacle to utilizing biochemical and statistical inference networks is the relative incompleteness of constructed networks. Genome-scale network reconstructions for humans are emerging in recent years (De Smet & Marchal, 2010; Duarte et al., 2007; Hyduke & Palsson, 2010; Jerby et al., 2010; Ma et al., 2007; Oberhardt et al., 2009; Rhodes et al., 2005; Vo et al., 2004; Zhao et al., 2010), with important implications for the integrated analyses of increasingly abundant genome sequences and transcriptomics data (see Figure 1). In the absence of more complete experimentally determined networks, there are multiple statistically inferred network configurations that would arrive at comparable molecular outputs, a fact that renders brute force trial and error computationally prohibitive and requires methods to avoid significant over fitting concerns. Despite these current limitations, significant efforts have been devoted to facilitate construction of comprehensive networks in cancer (Edelman et al., 2010). Improving the quality of network information is essential to understanding the complex ways in which cells go awry in cancer. As might be expected, many sets of interacting genes are well conserved across organisms. Examining conserved interactions across other eukaryotes has boosted the construction of human protein-protein interaction networks (Huang et al., 2007; Rhodes et al., 2005). Remarkably, mutations within these interacting gene sets can result in dramatically different phenotypes (McGary et al., 2010). For example, reduced growth in yeast deletion strains under particular conditions parallels abnormal angiogenesis phenotype in mice, Xenopus and human. These analogous phenotypes, or phenologs, can help to identify gene-phenotype associations across organisms. Examining phenolog-associated subnetworks resulted in discovery of biologically relevant molecular players at a rate 34 times higher than expected, given the annotation rate (McGary et al., 2010). Identifying phenologs helps to expand the information base from which non-intuitive associations with possible relevance to cancer can be discovered.
Figure 1.
Human-associated genome-scale information sources available.
Integrating heterogeneous networks can synergistically improve the accuracy and informative value of networks. For example, incorporating additional measurements of regulation, such as transcription factor binding (Chromatin Immunoprecipitation or ChIP-seq) and non-coding RNA regulation will enable a more comprehensive model of regulatory control (Qiu et al., 2010; Wang et al.). Furthermore, integrating a transcriptional regulatory network with a metabolic network substantially increases the predictive accuracy of the metabolic states in microbial systems under multiple conditions (Chandrasekaran & Price, 2010), and can be leveraged to aid more accurate network reconstructions for cell and tissue types in humans. Such capabilities need to be extended to human cells, which will provide increasing power to put observed molecular diagnostic changes in biological context—ultimately leading to better coupling between molecular diagnostics and therapy design.
Context-based diagnostics
Networks provide biological context for gene expression that aids a more comprehensive and accurate reconstruction of cellular systems. However, clinical practicality favors small sets of diagnostic markers, which renders exhaustive characterization of network state infeasible for individual diagnosis. A potential resolution involves isolating small sets of easily detectable markers whose behavior is most indicative of relevant subnetwork state. The collective value of these context-dependent markers would be their ability to identify the disease state within a system better than individual markers. The coupling of such context-dependent marker sets has been applied to assessing treatment strategies in breast cancer (Jordan, 1993). As mentioned previously, ER is a diagnostic marker for breast cancer that also is predictive of the effectiveness of endocrine therapy. Additionally, progesterone receptor (PR) expression levels in the context of ER provides significantly better predictions of therapy response than either PR or ER alone (Osborne et al., 2005). Identifying context dependent markers like ER and PR in collective panels would help to better account for the many possible low-penetrance alleles and variants believed to underlie various cancers. Utilizing diagnostic panels of these context-dependent markers leverages the clearer reconstruction of cellular states provided by network integration, thereby posing the potential for forming informative and easily interpretable markers for personalized medicine.
CONCLUSION
Prior to the first full sequencing of the human genome, it was estimated that humans had between 50,000 and 100,000 genes. We now know there to be around 20,000 genes. Sequencing of the human genome and follow-up studies revealed unanticipated genomic complexity within our cellular systems. Although we found significantly fewer genes than expected, the regulation of those genes has turned out to be far more intricate than anticipated. Like the process of elucidating the complexity of the human genome, the search for molecular cancer diagnostics will likely result in similar surprises.
As we continue to grapple with the many levels of biological complexity, our ability to construct better representations of cellular systems will depend on more accurate and comprehensive measurement techniques. Our analytical efforts to build better predictive models that isolate the most meaningful diagnostic molecular markers will undoubtedly also improve. Collectively, advances in these areas will result in unforeseen levels of complexity, all of which will reflect the biological and physiological complexity of the body. Our best test for how well we comprehend this complexity will be in our ability to detect, predict and ultimately treat disease.
In its own way, human biology has attempted to tackle the same challenges of molecular cancer diagnostics. Biology’s answer is the immune system. Like molecular diagnostics, the immune system seeks to identify malignant perturbations as early as possible, in large part, by relying on targeted specificity. Though biology has informed our approach to molecular diagnostic approaches, as it has done for numerous other technological advances, we must improve upon existing biology if we are to succeed. In some ways, molecular cancer diagnostics can be seen as the next evolutionary step of our immune system. Though improving upon the immune system is certainly daunting, systems biology provides us with an unprecedented bird’s eye view of cellular system organization, which can provide the necessary context to accomplish this task. The present rate of progress should give us confidence in our ability to successfully meet the very significant challenges that remain.
Acknowledgments
The authors gratefully acknowledge funding from the NIH-NCI Howard Temin Pathway to Independence Award in Cancer Research, the Grand Duchy of Luxembourg-Institute for Systems Biology Consortium, a Young Investigator Grant from the Roy J. Carver Charitable Trust and a National Science Foundation Graduate Research Fellowship (SM).
References
- AACC. Tumor Markers. 2010 Available online at: http://labtestsonline.org/understanding/analytes/tumor_markers/glance-3.html.
- Adkins Jn, Varnum Sm, Auberry Kj, Moore Rj, Angell Nh, Smith Rd, Springer Dl, Pounds Jg. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1(12):947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup Db, Wilhite Se, Ledoux P, Rudnev D, Evangelista C, Kim If, Soboleva A, Tomashevsky M, Marshall Ka, Phillippy Kh, Sherman Pm, Muertter Rn, Edgar R. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37(Database issue):D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hur A, Ong Cs, Sonnenburg S, Scholkopf B, Ratsch G. Support vector machines and kernels for computational biology. PLoS Comput Biol. 2008;4(10):e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild Ah, Yao G, Chang Jt, Wang Q, Potti A, Chasse D, Joshi Mb, Harpole D, Lancaster Jm, Berchuck A, Olson Ja, Jr, Marks, Dressman Hk, West M, Nevins Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Burchard J, Zhang C, Liu Am, Poon Rt, Lee Np, Wong Kf, Sham Pc, Lam By, Ferguson Md, Tokiwa G, Smith R, Leeson B, Beard R, Lamb, Lim L, Mao M, Dai H, Luk Jm. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Price Nd. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107(41):17845–17850. doi: 10.1073/pnas.1005139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Hy, Lee E, Liu Yt, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin Mf, Kellis M, Lindblad-Toh K, Lander Es. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci U S A. 2007;104(49):19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane Ac, Howlin J. Molecular profiling of breast cancer: transcriptomic studies and beyond. Cell Mol Life Sci. 2007;64(24):3185–3200. doi: 10.1007/s00018-007-7387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dati C, Antoniotti S, Taverna D, Perroteau I, De Bortoli M. Inhibition of c-erbB-2 oncogene expression by estrogens in human breast cancer cells. Oncogene. 1990;5(7):1001–1006. [PubMed] [Google Scholar]
- De La Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4(10):769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- De Smet R, Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol. 2010;8(10):717–729. doi: 10.1038/nrmicro2419. [DOI] [PubMed] [Google Scholar]
- Duarte Nc, Becker Sa, Jamshidi N, Thiele I, Mo Ml, Vo Td, Srivas R, Palsson Bo. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104(6):1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin Rm, Abecasis Gr, Altshuler Dl, Auton A, Brooks Ld, Gibbs Ra, Hurles Me, Mcvean Ga. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy Ja, Hood L, Price Nd, Geman D. Identifying tightly regulated and variably expressed networks by Differential Rank Conservation (DIRAC) PLoS Comput Biol. 2010;6(5):e1000792. doi: 10.1371/journal.pcbi.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman Lb, Eddy Ja, Price Nd. In silico models of cancer. Wiley Interdiscip Rev Syst Biol Med. 2010;2(4):438–459. doi: 10.1002/wsbm.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards Bk, Ward E, Kohler Ba, Eheman C, Zauber Ag, Anderson Rn, Jemal A, Schymura Mj, Lansdorp-Vogelaar I, Seeff Lc, Van Ballegooijen M, Goede Sl, Ries Lag. Annual Report to the Nation on the Status of Cancer, 1975–2006, Featuring Colorectal Cancer Trends and Impact of Interventions (Risk Factors, Screening, and Treatment) to Reduce Future Rates. Cancer. 2010;116(3):544–573. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn Pg, Baylin Sb, Herman Jg. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- Fletcher O, Houlston Rs. Architecture of inherited susceptibility to common cancer. Nat Rev Cancer. 2010;10(5):353–361. doi: 10.1038/nrc2840. [DOI] [PubMed] [Google Scholar]
- Fodde R, Smits R. Cancer biology. A matter of dosage. Science. 2002;298(5594):761–763. doi: 10.1126/science.1077707. [DOI] [PubMed] [Google Scholar]
- Gandellini P, Folini M, Zaffaroni N. Emerging role of microRNAs in prostate cancer: implications for personalized medicine. Discov Med. 2010;9(46):212–218. [PubMed] [Google Scholar]
- Gatza Ml, Lucas Je, Barry Wt, Kim Jw, Wang Q, Crawford Md, Datto Mb, Kelley M, Mathey-Prevot B, Potti A, Nevins A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107(15):6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geman D, D’avignon C, Naiman Dq, Winslow Rl. Classifying gene expression profiles from pairwise mRNA comparisons. Stat Appl Genet Mol Biol. 2004;3:Article19. doi: 10.2202/1544-6115.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochola Lf, Zeron-Medina J, Meriaux S, Bond Gl. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol. 2010;2(5):a001032. doi: 10.1101/cshperspect.a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Ra, Shah N, Wang Kc, Kim J, Horlings Hm, Wong Dj, Tsai Mc, Hung T, Argani P, Rinn Jl, Wang Y, Brzoska P, Kong B, Li R, West Rb, Van De Vijver Mj, Sukumar S, Chang Hy. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris Ra, Wang T, Coarfa C, Nagarajan Rp, Hong C, Downey Sl, Johnson Be, Fouse Sd, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg Kj, Gu J, Echipare L, O’geen H, Lister R, Pelizzola M, Xi Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi Op, Wilfond B, Borg A, Trent J, Raffeld M, Yakhini Z, Ben-Dor A, Dougherty E, Kononen J, Bubendorf L, Fehrle W, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344(8):539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Hicks Dg, Tubbs Rr. Assessment of the HER2 status in breast cancer by fluorescence in situ hybridization: a technical review with interpretive guidelines. Hum Pathol. 2005;36(3):250–261. doi: 10.1016/j.humpath.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Hood L, Heath, Phelps Me, Lin By. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306(5296):640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- Huang H, Liu Cc, Zhou Xj. Bayesian approach to transforming public gene expression repositories into disease diagnosis databases. Proc Natl Acad Sci U S A. 2010;107(15):6823–6828. doi: 10.1073/pnas.0912043107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Tw, Lin Cy, Kao Cy. Reconstruction of human protein interolog network using evolutionary conserved network. BMC Bioinformatics. 2007;8:152. doi: 10.1186/1471-2105-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol Mj, Kenzelmann-Broz D, Khalil Am, Zuk O, Amit I, Rabani M, Attardi Ld, Regev A, Lander Es, Jacks T, Rinn Jl. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D, Lee Iy, Yoo H, Gehlenborg N, Cho Jh, Petritis B, Baxter D, Pitstick R, Young R, Spicer D, Price Nd, Hohmann Jg, Dearmond Sj, Carlson Ga, Hood Le. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyduke Dr, Palsson Bo. Towards genome-scale signalling-network reconstructions. Nat Rev Genet. 2010;11(4):297–307. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- Ibrahim Jg, Chu H, Chen Lm. Basic concepts and methods for joint models of longitudinal and survival data. J Clin Oncol. 2010;28(16):2796–2801. doi: 10.1200/JCO.2009.25.0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry Ra, Warren D, Spencer F, Kim If, Biswal S, Frank Bc, Gabrielson E, Garcia Jg, Geoghegan J, Germino G, Griffin C, Hilmer Sc, Hoffman E, Jedlicka Ae, Kawasaki E, Martinez-Murillo F, Morsberger L, Lee H, Petersen D, Quackenbush J, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2(5):345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol. 2010;6:401. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones Pa, Baylin Sb. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones Rl, Salter J, A’hern R, Nerurkar A, Parton M, Reis-Filho Js, Smith Ie, Dowsett M. Relationship between oestrogen receptor status and proliferation in predicting response and long-term outcome to neoadjuvant chemotherapy for breast cancer. Breast Cancer Res Treat. 2010;119(2):315–323. doi: 10.1007/s10549-009-0329-x. [DOI] [PubMed] [Google Scholar]
- Jordan Vc. Fourteenth Gaddum Memorial Lecture. A current view of tamoxifen for the treatment and prevention of breast cancer. Br J Pharmacol. 1993;110(2):507–517. doi: 10.1111/j.1476-5381.1993.tb13840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson Ag., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal P, Tan Lk, Chen B. Correlation of HER-2 status with estrogen and progesterone receptors and histologic features in 3,655 invasive breast carcinomas. Am J Clin Pathol. 2005;123(4):541–546. doi: 10.1309/YMJ3-A83T-B39M-RUT9. [DOI] [PubMed] [Google Scholar]
- Lander Es, Linton Lm, Birren B, Nusbaum C, Zody Mc, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, Levine R, Mcewan P, Mckernan K, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lapidus Rg, Nass Sj, Davidson Ne. The loss of estrogen and progesterone receptor gene expression in human breast cancer. J Mammary Gland Biol Neoplasia. 1998;3(1):85–94. doi: 10.1023/a:1018778403001. [DOI] [PubMed] [Google Scholar]
- Leidinger P, Keller A, Heisel S, Ludwig N, Rheinheimer S, Klein V, Andres C, Staratschek-Jox A, Wolf J, Stoelben E, Stephan B, Stehle I, Hamacher J, Huwer H, Lenhof Hp, Meese E. Identification of lung cancer with high sensitivity and specificity by blood testing. Respir Res. 2010;11:18. doi: 10.1186/1465-9921-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch Kp. Gene-environment interaction and the genetics of depression. J Psychiatry Neurosci. 2004;29(3):174–184. [PMC free article] [PubMed] [Google Scholar]
- Li J, Lenferink Ae, Deng Y, Collins C, Cui Q, Purisima Eo, O’connor-Mccourt Md, Wang E. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 2010;1(4):1–8. doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Jj, Cutler G, Li W, Pan Z, Peng S, Hoey T, Chen L, Ling Xb. Multiclass cancer classification and biomarker discovery using GA-based algorithms. Bioinformatics. 2005;21(11):2691–2697. doi: 10.1093/bioinformatics/bti419. [DOI] [PubMed] [Google Scholar]
- Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni Jc, Mason Ce, Mane Sm, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Kj, Fournier Mv, Reddy Gp, Pardee Ab. A need for basic research on fluid-based early detection biomarkers. Cancer Res. 2010;70(13):5203–5206. doi: 10.1158/0008-5472.CAN-10-0987. [DOI] [PubMed] [Google Scholar]
- Mcgary Kl, Park Tj, Woods Jo, Cha Hj, Wallingford Jb, Marcotte Em. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107(14):6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell Ps, Parkin Rk, Kroh Em, Fritz Br, Wyman Sk, Pogosova-Agadjanyan El, Peterson A, Noteboom J, O’briant Kc, Allen A, Lin Dw, Urban N, Drescher Cw, Knudsen Bs, Stirewalt Dl, Gentleman R, Vessella Rl, Nelson Ps, Martin Db, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore Mj, Wang Q, Kennedy Cj, Silver Pa. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142(4):625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouridsen Ht, Rose C, Brodie Ah, Smith Ie. Challenges in the endocrine management of breast cancer. Breast. 2003;12 (Suppl 2):S2–19. doi: 10.1016/s0960-9776(03)80158-3. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Gonzalgo Ml, Yegnasubramanian S, Lin X, De Marzo Am, Nelson Wg. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem. 2004;91(3):540–552. doi: 10.1002/jcb.10740. [DOI] [PubMed] [Google Scholar]
- Oberhardt Ma, Palsson Bo, Papin Ja. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne Ck. Steroid hormone receptors in breast cancer management. Breast Cancer Res Treat. 1998;51(3):227–238. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- Osborne Ck, Schiff R, Arpino G, Lee As, Hilsenbeck Vg. Endocrine responsiveness: understanding how progesterone receptor can be used to select endocrine therapy. Breast. 2005;14(6):458–465. doi: 10.1016/j.breast.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Parsons Dw, Jones S, Zhang X, Lin Jc, Leary Rj, Angenendt P, Mankoo P, Carter H, Siu Im, Gallia Gl, Olivi A, Mclendon R, Rasheed Ba, Keir S, Nikolskaya T, Nikolsky Y, Busam Da, Tekleab H, Diaz La, Jr, Hartigan J, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou Cm, Sorlie T, Eisen Mb, Van De Rijn M, Jeffrey Ss, Rees Ca, Pollack, Ross Dt, Johnsen H, Akslen La, Fluge O, Pergamenschikov A, Williams C, Zhu Sx, Lonning Pe, Borresen-Dale Al, Brown Po, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pomraning Kr, Smith Km, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47(3):142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Price Nd, Shmulevich I. Biochemical and statistical network models for systems biology. Curr Opin Biotechnol. 2007;18(4):365–370. doi: 10.1016/j.copbio.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price Nd, Trent J, El-Naggar Ak, Cogdell D, Taylor E, Hunt Kk, Pollock Re, Hood L, Shmulevich I, Zhang W. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad Sci U S A. 2007;104(9):3414–3419. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Wang J, Yao P, Wang E, Cui Q. microRNA evolution in a human transcription factor and microRNA regulatory network. BMC Syst Biol. 2010;4:90. doi: 10.1186/1752-0509-4-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser Jm, O’shea Ek. Noise in gene expression: origins, consequences, and control. Science. 2005;309(5743):2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendis-Antonio O, Checa A, Encarnacion S. Modeling core metabolism in cancer cells: surveying the topology underlying the Warburg effect. PLoS One. 2010;5(8):e12383. doi: 10.1371/journal.pone.0012383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes Dr, Tomlins Sa, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan Am. Probabilistic model of the human protein-protein interaction network. Nat Biotechnol. 2005;23(8):951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Richon Vm, Sandhoff Tw, Rifkind Ra, Marks Pa. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A. 2000;97(18):10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan Bm, Robles Ai, Harris Cc. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf Hj, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis Rw, Brown Po. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid Lh, Jones Wd, Shippy R, Warrington Ja, Baker Sc, Collins Pj, De Longueville F, Kawasaki Es, Lee Ky, Luo Y, Sun Ya, Willey Jc, Setterquist Ra, Fischer Gm, Tong W, Dragan Yp, Dix Dj, Frueh Fw, Goodsaid Fm, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood Ld, Parsons Dw, Lin J, Barber Td, Mandelker D, Leary Rj, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz Sd, Willis J, Dawson D, Willson Jk, Gazdar Af, Hartigan J, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Slamon Dj, Clark Gm, Wong Sg, Levin Wj, Ullrich A, Mcguire Wl. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Slamon Dj, Godolphin W, Jones La, Holt Ja, Wong Sg, Keith De, Levin Wj, Stuart Sg, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Slatter Jg, Templeton Ie, Castle Jc, Kulkarni A, Rushmore Th, Richards K, He Y, Dai X, Cheng Oj, Caguyong M, Ulrich Rg. Compendium of gene expression profiles comprising a baseline model of the human liver drug metabolism transcriptome. Xenobiotica. 2006;36(10–11):938–962. doi: 10.1080/00498250600861728. [DOI] [PubMed] [Google Scholar]
- Smith Ds, Catalona Wj. Rate of change in serum prostate specific antigen levels as a method for prostate cancer detection. J Urol. 1994;152(4):1163–1167. doi: 10.1016/s0022-5347(17)32528-4. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou Cm, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen Mb, Van De Rijn M, Jeffrey Ss, Thorsen T, Quist H, Matese Jc, Brown Po, Botstein D, Eystein Lonning P, Borresen-Dale Al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron Js, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou Cm, Lonning Pe, Brown Po, Borresen-Dale Al, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E, Kittrell F, Medina D, Berget Sm. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18(24):3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- Stratton Mr, Campbell Pj, Futreal Pa. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam Ds, Isaacs C. Utilizing prognostic and predictive factors in breast cancer. Curr Treat Options Oncol. 2005;6(2):147–159. doi: 10.1007/s11864-005-0022-1. [DOI] [PubMed] [Google Scholar]
- Taylor Iw, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana Jl. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27(2):199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- Van De Vijver Mj, He Yd, Van’t Veer Lj, Dai H, Hart Aa, Voskuil Dw, Schreiber Gj, Peterse Jl, Roberts C, Marton Mj, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, Van Der Velde T, Bartelink H, Rodenhuis S, Rutgers Et, Friend Sh, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Van Ravenzwaay B, Cunha Gc, Leibold E, Looser R, Mellert W, Prokoudine A, Walk T, Wiemer J. The use of metabolomics for the discovery of new biomarkers of effect. Toxicol Lett. 2007;172(1–2):21–28. doi: 10.1016/j.toxlet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz Ja. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Venables Jp, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, Gendron D, Brosseau Jp, Thibault P, Lucier Jf, Tremblay K, Prinos P, Wellinger Rj, Chabot B, Rancourt C, Elela Sa. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68(22):9525–9531. doi: 10.1158/0008-5472.CAN-08-1769. [DOI] [PubMed] [Google Scholar]
- Venter Jc, Adams Md, Myers Ew, Li Pw, Mural Rj, Sutton Gg, Smith Ho, Yandell M, Evans Ca, Holt Ra, Gocayne Jd, Amanatides P, Ballew Rm, Huson Dh, Wortman, Zhang Q, Kodira Cd, Zheng Xh, Chen L, Skupski M, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl Hc, Matarese F, Marks H, Denissov S, Butter F, Lee Kk, Olsen Jv, Hyman Aa, Stunnenberg Hg, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Vo Td, Greenberg Hj, Palsson Bo. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J Biol Chem. 2004;279(38):39532–39540. doi: 10.1074/jbc.M403782200. [DOI] [PubMed] [Google Scholar]
- Vogel C, Abreu Rde S, Ko D, Le Sy, Shapiro Ba, Burns Sc, Sandhu D, Boutz Dr, Marcotte Em, Penalva Lo. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Eschenbach Ac. A vision for the National Cancer Program in the United States. Nat Rev Cancer. 2004;4(10):820–828. doi: 10.1038/nrc1458. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang Y, Teng M, Zhang D, Li L, Liu Y. Signal transducers and activators of transcription-1 (STAT1) regulates microRNA transcription in interferon gamma-stimulated HeLa cells. PLoS One. 2010;5(7):e11794. doi: 10.1371/journal.pone.0011794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Fact Sheet: The top 10 causes of death. 2008 Available online at: http://www.who.int/mediacentre/factsheets/fs310/en/index.html.
- Winrow Cj, Tanis Kq, Rigby Am, Taylor Rr, Serikawa K, Mcwhorter M, Tokiwa Gy, Marton Mj, Stone Dj, Koblan Ks, Renger Jj. Refined anatomical isolation of functional sleep circuits exhibits distinctive regional and circadian gene transcriptional profiles. Brain Res. 2009;1271:1–17. doi: 10.1016/j.brainres.2009.02.083. [DOI] [PubMed] [Google Scholar]
- Zhao J, Geng C, Tao L, Zhang D, Jiang Y, Tang K, Zhu R, Yu H, Zhang W, He F, Li Y, Cao Z. Reconstruction and analysis of human liver-specific metabolic network based on CNHLPP data. J Proteome Res. 2010;9(4):1648–1658. doi: 10.1021/pr9006188. [DOI] [PubMed] [Google Scholar]