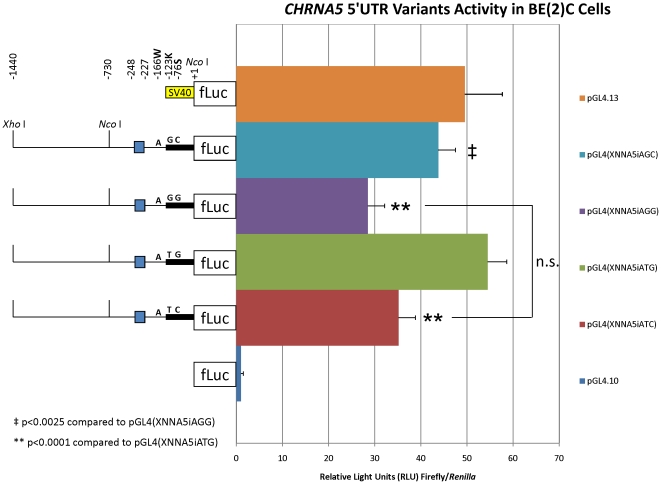
Figure 3. Relative CHRNA5 5′UTR variant luciferase activity in BE(2)-C cells.
Shown are the averages ± standard deviations of four independent experiments each done in triplicate. The SV40 promoter of the positive control vector pGL4.13[luc2/SV40] is indicated by a yellow box. The “promoterless” negative (background) control vector pGL4.10[luc2] is shown without a promoter region in front of the firefly luciferase (“fLuc”) reporter gene. For experimental CHRNA5 promoter-5′UTR constructs, thin and thick black lines represent the promoter and 5′UTR portion of the construct, respectively. Positions of the Xho I (−1440) and Nco I (+1) sites used for subcloning are indicated, as is an internal Nco I site (−730). The positions and identities of the rs3841324 (−227 to −248, blue box indicates the normal or “i” allele), rs503464 (−166W), rs55853698 (−123K) and rs55781567 (−76S) polymorphisms within each construct are indicated. Note that the “A” allele at rs503464 (−166 W) was not changed to “T” (as defined by haplotypes in Europeans, [26]) in the pGL(XNNA5iAGG) construct, in order to isolate the 5′UTR SNPs for effects on translation in an equivalent promoter context for each construct. ‡p<0.0025 compared to pGL4(XNNA5iAGG) and **p<0.0001 compared to pGL4(XNNA5iATG) by ANOVA with Tukey's HSD post-hoc analysis. “n.s.” indicates not significant.