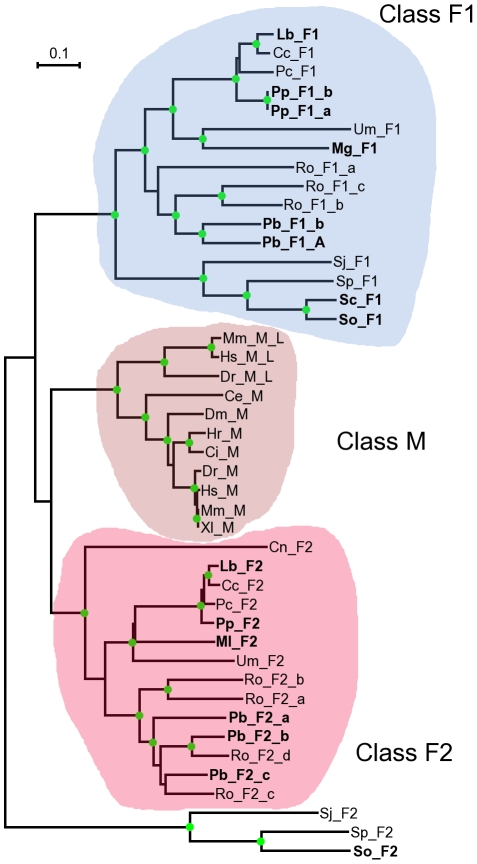
Figure 1. Phylogenetic distribution of CSL proteins used in this study.
An unrooted neighbour-joining phylogenetic tree of all CSL proteins analysed in this study. Novel CSL sequences (labelled in bold) follow the taxonomical distribution of those published previously [13]. Schizosaccharomyces pombe (Sp), S. octosporus (So), S. japonicus (Sj) and S. cryophilus (Sc) belong to Taphrinomycotina, the basal subphyllum of ascomycetes. Coprinus cinereus (Cc), Cryptococcus neoformans (Cn), Laccaria bicolor (Lb), Malassezia globosa (Mg), Melampsora laricis-populina (Ml), Phanerochaete chrysosporium (Pc), Ustilago maydis (Um) and Postia placenta (Pp) are basidiomycetes. Rhizopus oryzae (Ro) and Phycomyces blakesleeanus (Pb) are zygomycetes. Representative metazoan CSL sequences are from human (Hs), mouse (Mm), zebrafish (Dr), Xenopus laevis (Xl), Ciona intestinalis (Ci), Halocynthia roretzi (Hr), fruit fly (Dm) and Caenorhabditis elegans (Ce). Paralogs are denoted by letter suffixes (see Table S1 for more information). The three CSL classes are indicated by coloured background (F1 – blue; F2 – red, M – brown). The class F2 fission yeast branch position is of low confidence and therefore not shaded. Green circles at nodes indicate ≥90% bootstrap stability. The scale bar indicates the number of amino acid substitutions per site.