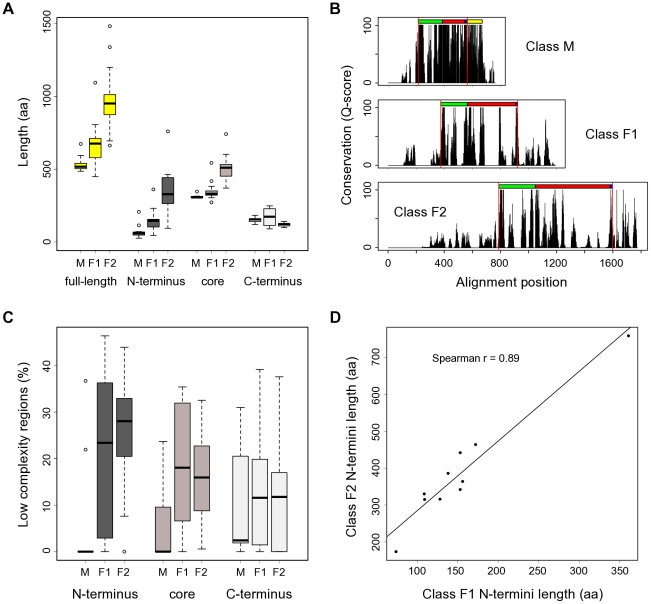
Figure 2. CSL protein length, organization and conservation.
(A) Fungal CSL proteins contain notable extensions in their N-termini (class F1, F2) and core (class F2). Whisker plots showing size distributions of the CSL proteins used in this study both for full-length sequences and their respective N-terminal, core, and C-terminal regions. M (n = 11), F1 (n = 16) and F2 (n = 17) denote the three distinct classes within the CSL family. (B) Sequence conservation profiles for the individual CSL classes (based on a gapped ClustalX protein alignment) show marked differences between the N-terminal, core, and C-terminal regions. The known domain composition is indicated above each profile: RHR-N (green), BTD (red), βC4 linker (blue), RHR-C (yellow; divergent in fungi). Red vertical lines show the partitioning into the 3 regions described in the main text. (C) Distribution of low-complexity regions across the CSL protein sequences and classes reveals a higher abundance of LCRs in the fungal homologs. The percentages of sequence scored as having a low complexity are shown. Note that the results for N-termini of class M are affected by the very short length of that region in this class. (D) The length ratios of N-termini between the F1 and F2 classes are conserved in all species tested. Each data point represents a fungal species; the coordinates are the corresponding class F1 and F2 paralog N-termini lengths, respectively. Mean values were plotted for species with multiple paralogs per class. Only species with both F1 and F2 representatives present in our dataset were included.