Figure 4.
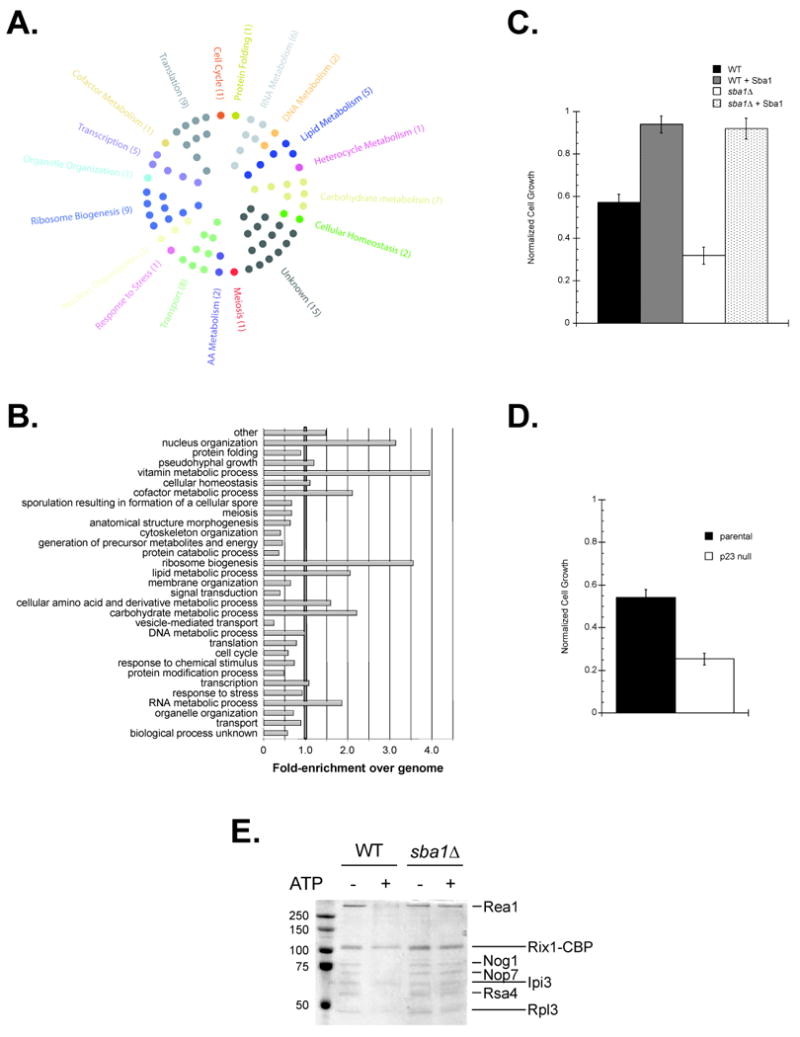
Sba1 physically interacts with proteins functioning in a wide variety of cellular process including ribosome biogenesis. Yeast ProtoArrays were used to detect proteins bound by Sba1. (A) Each hit was assigned to an initial cellular process using a GO Slim analysis and displayed or (B) all potential processes were considered for each gene and the enrichments in each category were determined relative to the distribution for all yeast ORFs. (C) The sensitivity of yeast to the ribosome inhibitor Hygromycin B (300 μg mL-1) fluctuates with Sba1 levels. (D) The Hygromycin B (100 μg mL-1) sensitivity of exponentially growing parental and p23 null MEFs was determined. Error bars represent the standard error of the mean. (E) Sba1 facilitates ATP-dependent release of ribosome biogenesis factors from Rix1-associated, nascent 60S particles. TAP purifications were performed using extracts prepared from wild type and sba1Δ yeast expressing Rix1-TAP, during the Calmodulin affinity step ATP (2 mM) was included, as indicated, and the EGTA eluates were analyzed by SDS-PAGE and Coomassie staining. The maturation factors known to dissociate in an ATP-dependent manner are demarked (short lines) along with the ATP-insensitive components (long lines) (Ulbrich et al., 2009).
