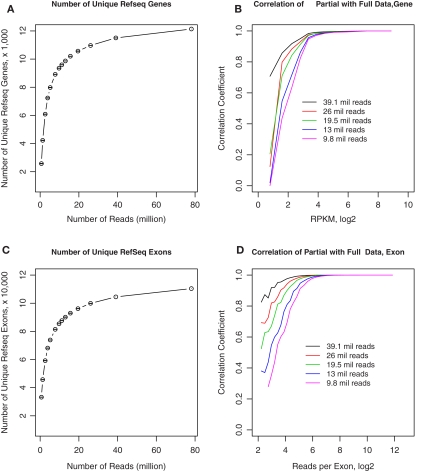
Figure 8.
Effect of sequencing depth on RNA-seq data quality. (A) We obtained 78 million 50-bp reads from rat 1. Different numbers of reads were randomly sampled from the full data set of 78 million reads. The number of unique RefSeq genes was calculated from the full and randomly sampled subsets. The total number of unique genes remained stable when greater than 9.8 million reads were analyzed. (B) Gene expression levels calculated from these randomly selected subsets were highly correlated with the full data set when RPKM is above ∼23, despite gradual decreasing the number of reads to 9.8 million. (C) The number of unique exons detected were calculated for the entire and randomly sampled subsets. The total number of exons remained stable when greater than 9.8 million reads were analyzed. (D) The number of reads for each exon obtained from the randomly sampled data sets were correlated to the entire data set. The correlation for exons with more than ∼25 reads was greater than 0.9 despite the decrease in reads to 9.8 million. At 19.5 million reads, exons with ∼8 reads still had correlation of ∼0.7 with values obtained from the entire data set. These results suggest that ∼15–20 million reads are sufficient to generate estimates of gene expression levels that are similar to those obtained from 70 to 80 million reads.