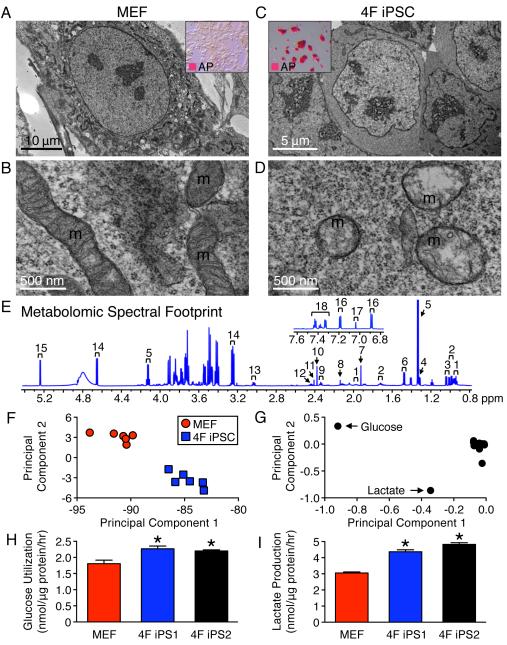
Figure 1. Nuclear reprogramming transforms mitochondrial structure inducing a distinct metabolomic footprint.
Nuclear reprogramming induced regression from elongated and cristae-rich mitochondria (m) of MEF (A, B) to spherical and cristae-poor remnant structures in four stemness factor derived iPSC (4F iPSC) (D). Insets demonstrate conversion of fibroblast monolayers (A) into compact clusters (C), which stained for the pluripotency marker AP. 1H NMR spectra of extracellular metabolites from 4F iPSC: 1 – isoleucine, 2 – leucine, 3 – valine, 4 – threonine, 5 – lactate, 6 – alanine, 7 – acetate, 8 – methionine, 9 – glutamate, 10 – pyruvate, 11 – succinate, 12 – glutamine, 13 – lysine, 14 – β-glucose, 15 – α-glucose, 16 – tyrosine, 17 – histidine and 18 – phenylalanine (E). Principal component analysis segregated 4F iPSC metabolomic phenotypes away from the MEF profile with principal component 1 accounting for 88% and 2 for 8% of the total variance (F). The loading plot assigned glucose and lactate as key metabolites contributing to segregation (G). Increased utilization of glucose and production of glycolytic end products in excess of MEF (H, I) were reproduced in independent iPSC lines (4F iPS1 and 4F iPS2). Values are mean ± SEM, n=6. * P<0.05 versus MEF. See also Figure S1.