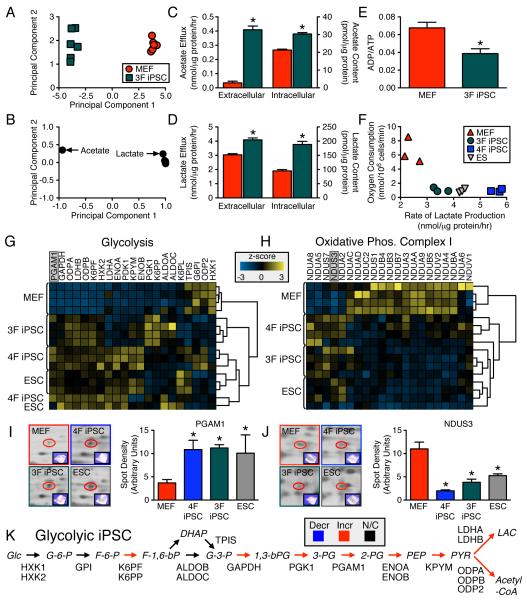
Figure 4. Metabolic reprogramming is independent of c-Myc transduction and supported by glycolytic/electron transport chain proteome switch.
1H NMR cellular metabolomic fingerprints (n=6) of three stemness factor induced iPSC (3F iPSC) segregated, away from the somatic, the acquired pattern (first principal component accounts for 97% and second for 1% of total variance) (A). Glycolytic end products, acetate and lactate, were key metabolites responsible for segregation (B). Intracellular content and efflux of acetate and lactate were significantly elevated in the 3F iPSC compared to MEF (C and D) and associated with reduced energy turnover (n=6) (E). Compared to MEF, 3F iPSC had lower maximal oxidative capacity and higher lactate production similar to that of ESC, albeit not fully overlapping with 4F iPSC (n=3) (F). Proteome-wide label-free quantification segregated iPSC away from MEF towards an ESC pattern based on agglomerative clustering of z-score transformed data (n=4) due to predominant glycolytic enzyme upregulation (G). Electron transport chain complex I subunits were predominantly downregulated in pluripotent cytotypes, which clustered away from MEF (H). 2-D gel quantification and MS/MS identification (n=3) independently confirmed glycolytic upregulation (I) and complex I downregulation (J). iPSC proteomic upregulation was mapped across the glycolytic pathway (K). Values are mean ± SEM. *P<0.05 versus MEF. Proteins are abbreviated by Swiss-Prot gene name. See also Figure S4 and Tables S1 and S2.