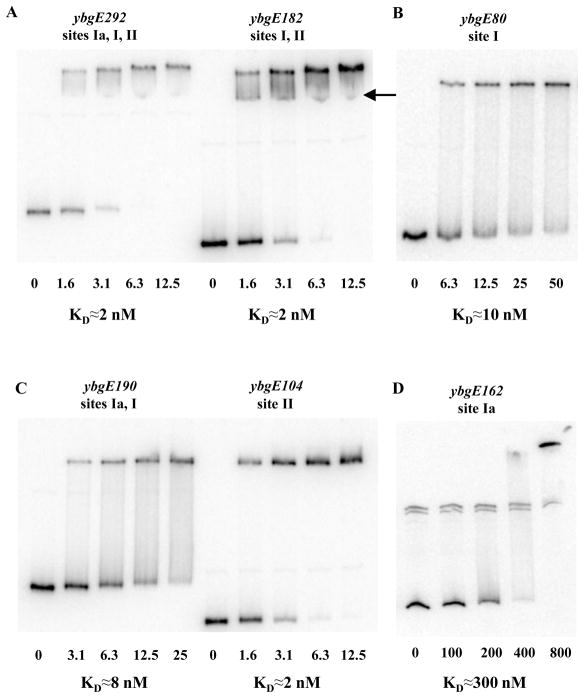
Fig. 4. Gel-shift assay of CodY affinity for ybgE DNA fragments.
Various labelled ybgE DNA fragments were incubated with increasing amounts of purified CodY in the presence of 10 mM ILV and 2 mM GTP. CodY concentrations used (nM of monomer) are indicated below each lane. The arrow indicates a likely complex in which CodY is bound only to one binding site as opposed to two sites simultaneously. KD, the apparent equilibrium dissociation constant, reflects the CodY concentration needed to shift 50% of DNA fragments under conditions ofvast CodY excess over DNA.