Abstract
An effective cellular delivery vector with enhanced intracellular retention was developed by conjugating a cell-penetrating peptide (CPP) with a fatty acid chain. The optimized lipopeptide (LP), myristoylated hendecaarginine (C14R11), penetrated cell membrane with high efficiency, and achieved superior metabolic stability and versatility as compared with unmodified oligoarginine CPPs, offering no adverse effect on cell viability and function. Cellular uptake, intracellular localization, cytotoxicity, and release kinetics of oligoarginines and LPs were investigated using flow cytometry analysis, cytotoxicity assay, and confocal microscopy. The cellular uptake efficiency and intracellular metabolic stability of C14R11 LP was further enhanced by replacing the L-arginine residues with D-arginine isomers. The cellular uptake and intracellular metabolic stability of D-form C14R11 (C14dR11) was significantly increased without any noticeable cytotoxicity compared to the unmodified parent hepta-arginine CPP or L-arginine LPs.
Introduction
It is well known that certain natural proteins could penetrate cell membranes directly, bypassing receptors or transporters.1–5 Interestingly, the cell penetrating capability of these proteins is largely imparted by relative short peptide sequences, termed cell-penetrating peptides (CPPs). The universal penetrating property of CPPs has been demonstrated useful in facilitating cellular uptake of various molecular cargoes, either through a covalent or non-covalent manner.6–8 Among reported CPPs, an arginine rich peptide derived from human immunodeficiency virus 1 (HIV-1) trans-activating transcriptional activator (Tat) has gained much attention due to its efficiency, simplicity and aqueous solubility.9 The functional domain on Tat protein to control translocation is a short 9-mer, RKKRRQRRR.1, 2, 5
It has been verified that the positively charged arginine residues in arginine-rich Tat peptide are crucial to its membrane penetrating capability,10 nonetheless, the positive charge is not the key factor, because the cell penetrating property was vanished when the positively charged arginine residues were replaced by other positively charged amino acids such as lysine, ornithine, histidine, or citrulline suggesting that the cationic guanidine moiety on the arginine side-chain plays a critical role on this unique membrane translocation property.11 Therefore, arginine-rich CPPs including oligoarginine peptides have been selected as vectors to transport cargo molecules, such as various drugs, biomolecules, nanoparticles, and imaging reporters, into cytoplasm and nucleus of cells for various biological applications based on their high cellular uptake efficiency.9, 12–14 The efficiency of cellular uptake of CPPs depends on the number of arginine residues and shows increased cellular uptake efficiency as the arginine sequences increased. However, membrane penetration and nuclear accumulation is lessened in short (< 5) or long (> 15) arginine peptide sequences.11
A major obstacle in developing potent therapeutic agents is their poor bioavailability which prevents them from entering the lipid bilayer of plasma membrane, resulting in a diminished therapeutic efficacy. In order to achieve improved delivery into cytoplasm or intracellular compartments, hydrophobic enrichment has been introduced into therapeutic agents.15 Similarly, fatty acid moieties that could advance cellular association with the hydrophobic cell membranes have been incorporated onto various CPPs, such as proline-rich, arginine-rich, or cationic peptides.16–20 Previously others and we have reported that the oligoarginine peptides modified with fatty acid moieties could enhance delivery efficiency.21, 22 Later on, the fatty acids conjugated polyarginine, labelled with near-infrared probe, showed potential for in vivo neuroimaging, because it could cross blood-brain barrier (BBB) and accumulate in the mouse brain.23 Furthermore, fluorescent dye labelled myristoylated polyarginine peptide probe was utilized to track migration and homing of dendritic cells in vivo using the whole mouse optical imaging system.24
Prior efforts have confirmed that oligoarginine sequences tethered with fatty acid moieties could be effective delivery vectors, because such modifications could enhance membrane association and, potentially, would lead to improved delivery of cargo molecules into intracellular compartment. Here we describe a systemic evaluation of various lipopeptides (LPs) by introducing different fatty acid moieties at the N-terminus of various oligoarginine peptides, focusing on cellular uptake, intracellular distribution, cell toxicity, and metabolic stability with extended retention in Jurkat cells.
Results
Characterization of oligoarginines and lipopeptides (LPs)
LPs were synthesized as previously described with minor modification.21 A series of fatty acid groups, including lauroyl (C12), myristoyl (C14), and palmitoyl (C16), were attached to the N-terminus of amidated oligoarginine peptides (R7, R9, R11, R13, and R15) to compare length effect of oligoarginine as well as lipophilic fatty acid chains. The LPs were labeled with FITC on their C-terminal lysine side chains to monitor the cellular uptake and intracellular distribution of LPs by fluorescence microscopy and flow cytometry. The molecular weight (Mw) of the prepared oligoarginine peptides and LPs was confirmed by MALDI mass spectrometry (Table 1).
Table 1.
Characterization of synthesized LPs
| Name | Sequencea | Mw [M+H]+ | MALDI-TOF |
|---|---|---|---|
| R7 | B(R)7K(FITC)-NH2 | 1699.9 | 1700.6 |
| R9 | B(R)9K(FITC)-NH2 | 2012.3 | 2011.9 |
|
| |||
| C12R7 | Lauroyl-B(R)7K(FITC)-NH2 | 1883.6 | 1882.8 |
| C12R9 | Lauroyl-B(R)9K(FITC)-NH2 | 2196.0 | 2195.7 |
| C12R11 | Lauroyl-B(R)11K(FITC)-NH2 | 2508.4 | 2507.8 |
| C12R13 | Lauroyl-B(R)13K(FITC)-NH2 | 2820.7 | 2819.4 |
| C12R15 | Lauroyl-B(R)15K(FITC)-NH2 | 3133.1 | 3132.9 |
|
| |||
| C14R7 | Myristoyl-B(R)7K(FITC)-NH2 | 1910.7 | 1910.1 |
| C14R9 | Myristoyl-B(R)9K(FITC)-NH2 | 2223.1 | 2224.2 |
| C14R11 | Myristoyl-B(R)11K(FITC)-NH2 | 2535.4 | 2534.6 |
| C14dR11 | Myristoyl-B(dR)11K(FITC)-NH2 | 2535.4 | 2536.2 |
| C14R13 | Myristoyl-B(R)13K(FITC)-NH2 | 2848.8 | 2848.9 |
| C14R15 | Myristoyl-B(R)15K(FITC)-NH2 | 3161.1 | 3160.1 |
|
| |||
| C16R7 | Palmitoyl-B(R)7K(FITC)-NH2 | 1938.8 | 1938.2 |
| C16R9 | Palmitoyl-B(R)9K(FITC)-NH2 | 2251.1 | 2250.4 |
| C16R11 | Palmitoyl-B(R)11K(FITC)-NH2 | 2563.5 | 2563.7 |
| C16R13 | Palmitoyl-B(R)13K(FITC)-NH2 | 2875.8 | 2874.8 |
| C16R15 | Palmitoyl-B(R)15K(FITC)-NH2 | 3188.2 | 3188.0 |
B: β-alanine, dR: D-arginine, FITC: fluorescein isothiocyanate
Quantification of cell-associated LPs by flow cytometry
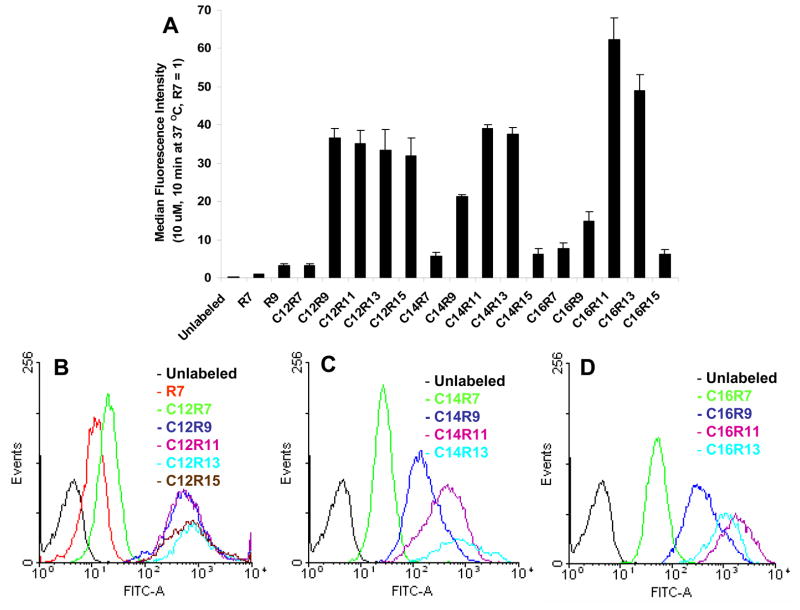
In order to evaluate the cellular uptake of LPs, Jurkat cells were incubated with LPs (10 μM) at 37 °C for 10 min, and then the fluorescence intensity was quantified by flow cytometry (Fig. 1). Hepta-arginine (R7) was chosen as a reference, because it was reported that R7 and Tat peptide have similar cell penetrating property.10 After 10 min incubation, high cellular association was observed with all lauroylated LPs, except C12R7 which only had a moderate gain in cellular uptake. Comparing to control R7, much higher fluorescent signals (30–40 folds, p < 0.01) were achieved with lauroylated LPs. The length of arginine peptide didn’t play a significant role at this condition (C12 series) (Fig. 1B). On the other hand, the fluorescence intensity of C14 and C16 LPs was elevated in an arginine length dependent manner till reaching continuum status. The fluorescence intensity reached maximal at C14R11 in C14 LPs (Fig. 1C) and C16R11 in C16 LPs (Fig. 1D). Further lengthening of the arginine residues to R15, however, decreased the cellular associating efficiency of LPs. In contrast, the effect of fatty acids is less clear at this time point (10 min) with flow cytometry analysis. Fluorescence intensity was only slightly increased by different fatty acid lengths in the R7 series with following order, C12R7 (3.24 ± 0.59) < C14R7 (5.68 ± 0.93) < C16R7 (7.76 ± 1.42). Similar trend was also observed in the R11 series, C12R11 (35.14 ± 3.49) < C14R11 (39.1 ± 0.92) < C16R11 (62.42 ± 5.54), and R13 series, C12R13 (33.44 ± 5.43) < C14R13 (37.67 ± 1.77) < C16R13 (49.05 ± 4.12). However, the patterns of fatty acid length effect in the R9 and R15 series were random, C12R9 (36.68 ± 2.47) > C14R9 (21.26 ± 0.61) > C16R9 (14.83 ± 2.59), and C12R15 (31.84 ± 4.86) ≫ C14R15 (6.3 ± 1.3) ≅ C16R15 (6.14 ± 1.3).
Fig. 1.
Cellular uptake of 10 μM LPs incubated with Jurkat cells for 10 min at 37 °C. (A) Jurkat cells incubated with LPs were washed, trypsin treated, PI stained, and analyzed by flow cytometry with 1–2 × 104 events. (B, C, and D) The histograms of C12, C14, or C16 LPs. Each experiment was performed at least three times in triplicate, and the results are expressed as the median change in fluorescence ± S.D. The median fluorescence intensity of R7 in Jurkat cell was set as 1.
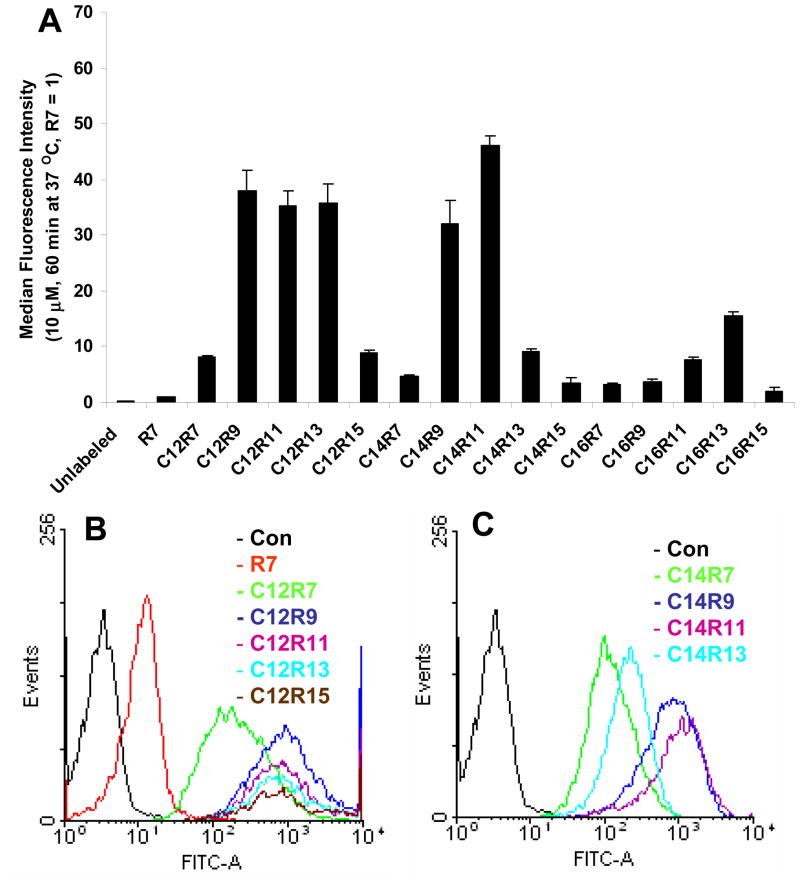
To further explore the effect of arginine residue and fatty acid moiety on cellular association between LPs and Jurkat cells, the incubation time was extended to 60 min (Fig. 2). The absolute median fluorescent intensity of R7 treated cells was increased from 16.2 ± 3.6 to 25.1 ± 2.1. For comparison purpose, the fluorescent signal of R7 incubated for 60 min was set as the reference. It was observed that the cellular uptake pattern of C12 LPs, except C12R15, at 60 min incubation was similar to that at 10 min (Fig. 1B vs. 2B). Although C14R9 and C14R11 LPs showed effective cellular association, C14R13 and C14R15 LPs only exhibited moderate cellular uptake (Fig. 2B). Surprisingly, the fluorescent signal intensity of C16 LPs was significantly reduced with extended incubation time, although they were still more efficient than R7. Taken together, relatively low cellular association was observed with long arginine residues (R13 and R15) and long fatty acids (C16) in the extended incubation time of Jurkat cells.
Fig. 2.
Cellular uptake of LPs in Jurkat cells incubated for 60 min. (A) Cells are incubated with 10 μM of LPs for 60 min at 37 °C. The fluorescence intensity of R7 at 60 min was set as 1. (B and C) The flow cytometry histograms of C12 and C14 LPs.
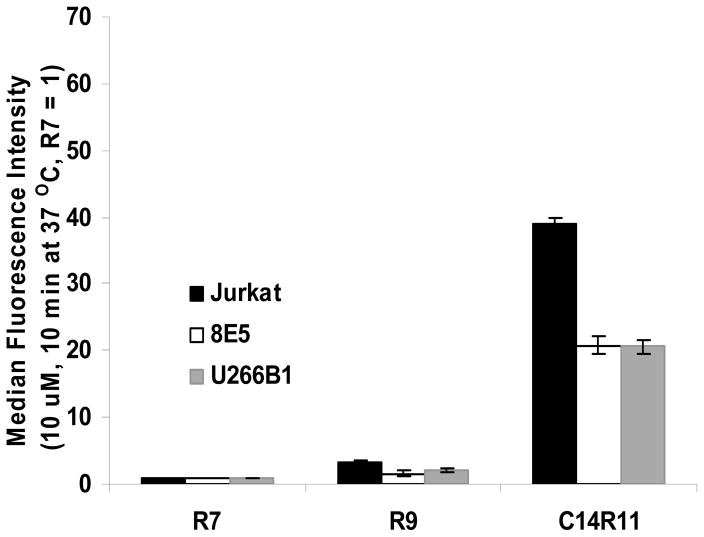
Cellular uptake efficiency was further evaluated with different cells. Jurkat cells, human T-lymphoma 8E5, and myeloma U266B1 cells were incubated with R7, R9 and C14R11. R9 was selected because of its derivatives, C12R9 and C14R9, showed significantly improved signal intensity compared to R7 containing LPs (C12R7 and C14R7) both in 10 and 60 min incubation (Fig. 1 and 2). Although R9 exhibited slightly higher cellular uptake than R7 peptide did, the signal intensity of R9 was far lower than it of acylated peptide, C14R11 (Fig. 3). 8E5 and U266B1 cells also showed similar cellular uptake trends with Jurkat cells.
Fig. 3.
Cellular uptake of 10 μM LPs incubated with various cell lines. Jurkat, 8E5, and U266B1 cells were incubated with R7, R9, and C14R11 LPs, washed, and analyzed by flow cytometry. The median fluorescence intensity of R7 in each cell line was set as 1.
Cell cytotoxicity
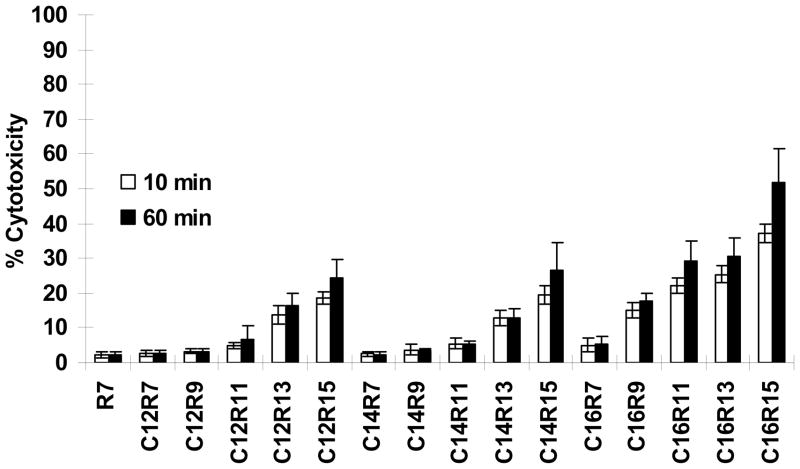
Cytotoxicity was determined by a conventional colorimetric method, in which the released lactate dehydrogenase (LDH) in culture medium, after exposure to LPs for 10 min or 60 min, converts the substrate tetrazolium salt into a red product with an affordable quantitative absorbance at 490 nm. R7 and its derivatives, including C12R7, C14R7, and C16R7 didn’t exhibit any significant toxicity to Jurkat cells. On the other hand, the cells treated with long arginine (R13 or R15) containing LPs had reduced viability in an arginine length-dependent manner, with R15 being more toxic than other oligoarginines on the cells (Fig. 4). In addition, all C16 LP series, except C16R7, caused significant LDH leakage. Although Jurkat cells treated with LPs for 60 min expressed higher cytotoxicity than 10 min treatment, the cytotoxicity difference between 10 min and 60 min incubation was not significant. The results suggest that LPs containing long chain fatty acid and/or long arginine residues could give a detrimental effect on cell viability. Therefore, LPs containing the long arginine chains (R13 and R15 LPs) and all C16 LPs were excluded from further study.
Fig. 4.
Effects of LPs on cell toxicity. Jurkat cells were incubated with 10μM of LPs for 10 min or 60 min at 37 °C. Data were normalized to the amount of maximum LDH released from the cultures treated with lysis solution alone and corrected for baseline LDH release from cultures exposed to buffer only.
Temperature dependency of cellular uptake capacity
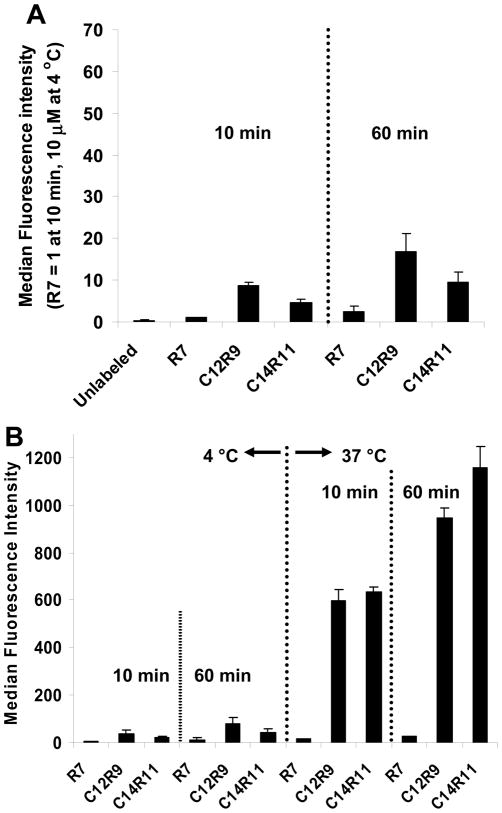
To investigate whether cellular uptake of LPs in cells is energy dependent, C12R9 and C14R11 LPs were selected for this study; while R7 was included as a control. Jurkat cells were incubated with LPs (10 μM) at 4 °C for 10 min or 60 min. Although extended incubation was not a crucial factor on cellular uptake of LPs at low temperature, signal intensities of C12R9 and C14R11 were increased with extended incubation (Fig. 5A). The cellular uptake of C12R9 was higher than C14R11 at 10 min and additive fluorescence intensity was observed in all peptides when the incubation time was extended to 60 min at 4 °C. However the total fluorescent intensity at 4 °C was much lower than it at 37 °C (Fig. 5B). It suggested that the uptake is most a temperature-dependent process, although a small fraction of the tested LPs including R7 were associated with cells effectively even at 4 °C.
Fig. 5.
Cellular uptake of LPs at different temperatures. (A) Relative fluorescent intensities of LPs. Cells were incubated with 10 μM of LPs (R7, C12R9, or C14R11) at 4 °C for 10–60 min. The fluorescence intensity of R7 incubated for 10 min at 4 °C was set as 1. (B) Absolute fluorescent intensity of cells at 4 °C or 37 °C for 10 or 60 min.
Intracellular distribution of LPs
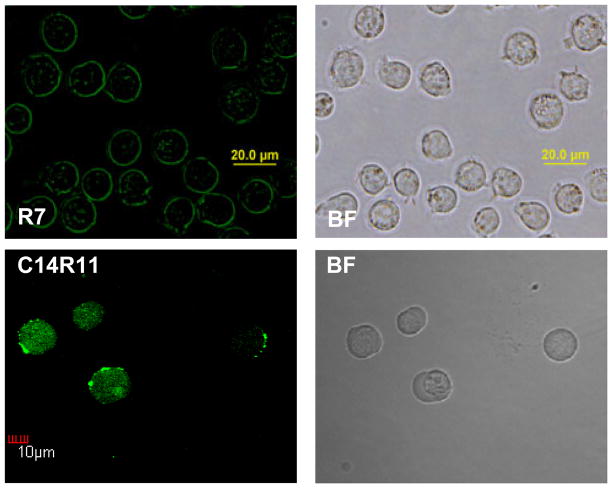
Since cell fixation procedure has been shown to cause serious artifacts in the intracellular localization of CPPs,25 all confocal laser microscopic observations were conducted in live cells. C14R11 was selected as a target delivery vector for its maximal cellular uptaking efficiency and low cytotoxicity. After 10 min of incubation with Jurkat cells, C14R11 was distributed both at the cytoplasm and plasma membrane of the cells (Fig. 6). On the other hand, dim spotty R7 fluorescence signal was seen inside of cells.
Fig. 6.
Intracellular distribution of LPs in Jurkat cells. Cells were treated with 10 μM of R7 and C14R11 for 10 min at 37 °C. The cell images were acquired by confocal microscopy.
LP stability in the Jurkat cells
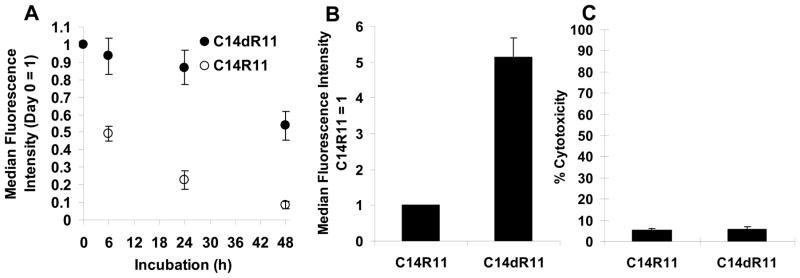
The intracellular fates of LPs were further studied by monitoring the fluorescence changes over a 48 h period. Surprisingly, in less than 6 h, 50 % of the fluorescence signal of C14R11 was lost, and only 9 % of signal was retained after 48 h. It was suspected that the instability of C14R11 is the key factor contributing to the loss of fluorescence signal. The oligoarginine peptide (R11) on C14R11 could be degraded by intracellular proteases, resulting in release of fluorochromes which were then pumped out of the cells. To improve intracellular metabolic stability, a D-arginine LP (C14dR11) was prepared and compared to C14R11. As expected, the flow cytometry data exhibited superior retention of C14dR11 signal intensity (Fig. 7A). Near 90 % and 54 % of fluorescence intensity were retained in the Jurkat cells at 24 h and 48 h, respectively. Moreover, the signal intensity of C14dR11 was 5-fold higher than that of C14R11 (Fig. 7B) and, importantly, C14dR11 did not show any serious cytotoxic effect on Jurkat cells (5.97 ± 0.9 %), either (Fig. 7C).
Fig. 7.
Comparison of C14dR11 or C14R11 on Jurkat cells. (A) Retention profiles of LPs. Cells were cultured with 10 μM of C14R11 or C14dR11 LPs at 37 °C for 10 min. Then cells were washed and reincubated in 6 well culture plates for a period of time (0, 6, 24, and 48 h) prior to flow cytometry analysis. (B) Cellular uptake of C14R11 or C14dR11 LPs. Cells was incubated with 10 μM of LPs at 37 °C for 10 min. The fluorescence intensity of C14R11 peptide was set as 1. (C) Cytotoxicity of C14R11 and C14dR11 LPs. Cells were incubated with 10 μM of LPs at 37 °C for 60 min. Experiments were performed in triplicate, mean ± SD.
Discussion
In the present study, we report a systemic approach to enhance cellular uptake efficiency of CPP into cells. Synthesized LPs have two functional units; the first unit is an oligoarginine peptide, which is known to penetrate cell membrane with high efficiency, and the other unit is a fatty acid moiety to improve cellular association.21 Cellular association of oligoarginine with cells was increased in moderate arginine residues (7 ≤ n ≤ 16),10, 11, 26–28 and our study also indicates that appropriate arginine lengths (9 ≤ n ≤ 13) could elevate cellular uptake compared to relatively short (R7) or long (R15) oligoarginine peptides (Fig. 1 and 2). Testing with different cell lines confirmed the general trend of labeling efficiency of the LPs (Fig. 3). Much higher cellular uptake was observed with C14R11 than that of R7 or R9 in all three tested cell lines.
It is well known that numerous natural proteins can be modified by covalently linked fatty acid moieties, usually myristoyl and palmitoyl groups, via an amide bond to the N-terminal amino group. The lipophilicity of proteins was enhanced after acylation, which is contributing to reversible association with membranes and controlling intracellular trafficking of proteins.29, 30 However, our prior study with a myristoylated random neutral peptide, BGFAGFAG, revealed that a myristoyl group alone was not enough to promote cellular uptake of the reference peptide.21 In a model system with phospholipid bilayer, the Kd of myristoylated peptide was calculated to be 10−4 M (8 Kcal/mol), which is too low to provide tight association with the plasma membrane of cell.31 In a separate model study, it was reported that a hydrophobic peptide modified by a single farnesyl group could not associate strongly with membrane, either.32 All those data imply that myristoylated peptide itself is not sufficient to provide effective cellular uptake, therefore, a second signal (‘two-signal hypotheses’) is required to achieve stable binding property of CPPs with cell membrane. Although either fatty acid (e.g. myristic acid) or basic peptide (e.g. oligoarginine) alone is not adequate for plasma membrane binding, if fatty acid and basic peptide are presented within the same protein or peptide template, the association with cell membrane could be enhanced. Previously, lipophilic moiety conjugated CPPs such as stearlylated arginine-rich peptides,17, 18, 33, 34 lipid conjugated cationic peptides,19, 20 or myristylated proline-rich peptides16 have been reported to achieve improved cellular uptake. Ruthenium-octaarginine conjugated with lipophilic fluorescence tag enhanced cellular uptake and intracellular localization of HeLa cells compared to Ru-octaarginine or Ru-fluorescence.35
R7 and its LPs (C12R7, C14R7, and C16R7) exhibited low level of LDH release, similar to Tat peptide, indicating that relatively short oligoarginine and its LPs are barely related with cytotoxicity.21, 28 However LPs consisted of long fatty acid chain and long oligoarginines showed significant cell toxicity (Fig. 4). Surprisingly, a dramatic improvement in cytotoxicity and cellular uptake efficiency was achieved when the palmitoyl group on C16R11 was replaced by a two-carbon shorter myristoyl group (C14R11), suggesting the critical contribution of lipophilic chains in cellular uptake and trafficking activity. This observation is partially consistent with a previous study in which the cellular delivery of cationic lipid peptide was enhanced with fatty acid lengths (C8 – C16), but cellular toxicity was also increased in parallel, indicating that the cellular trafficking of lipopeptides depends on a proper combination of arginines and fatty acid.19
Several studies have suggested that a major pathway for cellular translocation of cationic CPPs and arginine-rich peptides is via an energy dependent endocytosis,36–38 however, energy independent pathway has also been widely reported.13, 26, 35, 39. The signal intensity of LPs in our study was much reduced at 4 °C than it at 37 °C, suggesting the oligoarginines and LPs favor temperature dependent cellular uptake pathway, although a small fraction of LPs was still associated with cells at 4 °C (Fig. 5). It is possible that multiple pathways are involved in the cellular translocation and thus, an alternative pathway become active when one of them is blocked.40
C14dR11, in which all L-arginine residues were replaced with D-arginine residues, significantly increased cellular association with Jurkat cells compared with C14R11, which signal intensity was substantially decreased after 6 h and over 90 % of initial fluorescence signal was lost after two days. In contrast, C14dR11 offered 5-fold increased signal intensity and it has sustained for longer than 48 h (Fig. 7). It is suggesting that the improved cellular uptake of C14dR11 is mainly coming from its increased resistance to proteolysis.10, 17, 26
In this study, fluorescent signal was used as the solo indicator to validate the delivery efficiency of the prepared LPs. It is clear that the intracellular intensity of the fluorescence depends on several factors. Membrane penetrating efficiency is only one of them. Protease stability and intracellular retention also play important roles. By choosing appropriate arginine residue, peptide lengths and fatty acid chains, an optimized delivery vehicle could be crafted.
Up to date, CPPs have been widely applied as delivery vehicles transporting almost all kinds of payloads, such as small imaging radiotracers, drugs, nucleic acids, genes, proteins, and few hundred nanometer sized particles, into cells. Our results suggest that systematically optimized LPs are far better in cellular delivery than it of the unmodified CPPs. Consequently, the prepared D-form LPs could transport payloads in a great efficiency and could prolong intracellular accumulation of the payloads. Further demonstration of their potential usages is currently in progress.
Experimental
Peptide synthesis
The peptide sequences used are listed in Table 1. Peptides were synthesized by conventional Fmoc solid phase chemistry with β-alanine and lysine at either ends for conjugation with fatty acid and FITC moieties, respectively. 433A peptide synthesizer (Applied Biosystems, Foster City, CA) was loaded with Rink amide MBHA resin (0.25 mmol) and HBTU/HOBt as the activating and coupling reagents. Amino acids, Fmoc-Arg(Pbf)-OH, Fmoc-D-Arg(Pbf)-OH, Fmoc-Lys(Mtt)-OH, and Fmoc-β-Ala-OH, were from NovaBiochem (San Diego, CA). The Fmoc moiety was deprotected by 20% piperidine/DMF and the activation was catalyzed with NMP/DMF (0.4 M) for each cycle. D-form peptide was synthesized using the same protocol.
Fatty acid conjugated peptides
Peptide containing resin (0.08 mmol) was suspended in a freshly prepared DIEA/NMP solution (2 M, 5 ml). Fatty acylchloride (0.32 mmol; Sigma, St. Louis, MO) was added dropwisely into the resin slurry. The mixture was stirred gently at RT for 2 h. The product was collected by filtering through a vacuum manifold reservoir filter (Discovery Science, Deerfield, IL) and washed with NMP (5×) and methanol (5×).
Labeling with FITC
The protecting group 4-methyltrityl (Mtt) on the lysine side-chain was selectively removed with TFA in DCM (5 ml 1:99, v/v) for 2 min with stirring and repeated for 9 times following with DCM (5×), NMP (5×) and methanol (5×) washes. The free lysine-containing peptide was reacted with FITC (0.4 mmol, 155.8 mg; Sigma) in DIEA/DMF (1:4), and the slurry was stirred overnight at RT in dark.
Cleavage and purification
The bright yellow resin was washed with NMP and methanol. Cleavage cocktail (90:5:3:2 TFA/TA/EDT/anisole) applied to the resin for 3 h, the yellow peptide was precipitated by ether, and then purified by Vydac C18 reversed-phase HPLC (Varian, Palo Alto, CA) using 0.1 % TFA and acetonitrile as elution buffers to afford yellow cotton-like solid peptide after lyophilization. The Mw of prepared LPs was confirmed by AB4800 MALDI-TOF Analyzer (Applied Biosystems).
Cell cultures
Human Jurkat, human T-lymphoma 8E5, and human myeloma U266B1 cells (3.5 × 105 cells/ml), gifts from Texas Children’s Hospital, were cultured in RPMI-1640 supplemented with 10 % FBS and 1 % Penicillin/Streptomycin. Cells were grown on 75 cm2 culture flasks at 37 °C in a humidified 5 % CO2 incubator. The whole medium was changed every 2–3 days until cells reached near-confluency.
Flow cytometry analysis
The concentration of peptide solution was determined by absorption of FITC at 490 nm (ε = 67,000) in PBS buffer (pH = 7.2). Freshly prepared 100 μM of peptide stock solutions in PBS were added in each tube containing 3.5 × 105 of Jurkat, 8E5, or U266B1 cells in culture media to adjust 10 μM of final LP concentration. The cells were incubated for 10 or 60 min at 4 °C or 37 °C, followed by three washes with PBS. Cells were then incubated with 200 μl trypsin-EDTA for 5 min at 37 °C to remove the cell surface-bound peptides. At the end of trypsin treatment, 200 μl of culture medium was added. The cells were incubated with 200 μl ice-cold propidium iodide (1 μg/ml, Sigma) in PBS + 2 % FBS to assess cell viability. The cells were filtered with Cup Filcons cell strainer (70 μm). Cells staining with propidium iodide were excluded from the flow cytometry analysis. A total of 1–2 × 104 events were recorded using FACS LSR II (BD) and analyzed by WinMDI software (The Scripps Institute).
Cytotoxicity
The CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega, Madison, WI) was used to determine cytotoxic effects of LPs on Jurkat cells. The experiments were performed according to the instructions of the manufacturer. In order to avoid interference from phenol red, phenol red-free RPMI 1640 containing 10 % FBS was used in this assay. The 50 μl of culture suspension containing 2 × 104 cells was dispensed into 96 U-bottom well plates and the volume was adjusted to an appropriate concentration with a final volume of 100 μl by adding peptide solution. LP-induced toxicity was evaluated by measuring lactate dehydrogenase (LDH) activity released in the culture supernatant after 10 min or 60 min exposure to LPs at 37 °C. The plate was removed from the incubator and centrifuged at 250 × g for 4 min. The four controls listed below were performed with LPs assay. First, target cell spontaneous LDH release was included as a baseline control. Second, target cell maximum LDH release was carried out to determine 100% release of LDH. Next, volume correction control was used to correct for volume change caused by addition of lysis solution (10×). Finally, culture medium background was utilized to correct for LDH activity contributed by serum in culture medium. Supernatant (50 μl) from each well was transferred to a 96-well plate followed by addition of reconstituted substrate solution (50μl) and incubation of the reaction mixture at RT for 30 min, protected from light. Quantification of cell death was performed on a SpectraMax M2 Microplate (Molecular Devices, Sunnyvale, CA) at 490 nm and calculated using the following formula.
Fluorescence microscopy
Cells were treated with 10 μM of LPs for 10 min or 60 min at 37 °C, washed extensively with PBS, and cells were resuspended in PBS. Fluorescence image was captured by an Olympus Fluo View TM 1000 laser scanning confocal microscope (Center Valley, PA) using oil immersion 60× objective and glass-bottom culture slides with excitation by 488 nm (FITC-tagged LP) laser line. FV 1000 Viewer (Olympus) was used for image analysis.
Metabolic Stability of LPs on Jurkat cell
In order to evaluate metabolic stability of Jurkat cells induced by C14R11 or C14dR11 during cultures, 3.5 × 105 cells/ml were incubated in the absence (negative control) or presence of 10 μM LPs for 10 min at 37 °C. The cells were washed and cultured at 37 °C for 2 days with full culture media, A fraction of cells was harvested from culture dish at day 0, 1/4, 1, and 2 for flow cytometry analysis. A total of 2 × 104 events were recorded using BD FACS LSR II and analyzed by WinMDI.
Statistical analysis
The level of statistical comparison of the cellular uptake was evaluated by paired-sampleS t-test and the difference between control and experimental group was considered significant at p < 0.05.
Acknowledgments
This research work was supported in part by NIH CA135312, CA114149 and CA126752.
References
- 1.Frankel AD, Pabo CO. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 2.Green M, Loewenstein PM. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 3.Joliot AH, Triller A, Volovitch M, Pernelle C, Prochiantz A. New Biol. 1991;3:1121–1134. [PubMed] [Google Scholar]
- 4.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 5.Vives E, Brodin P, Lebleu B. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 6.Dietz GP, Bahr M. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Veldhoen S, Laufer SD, Trampe A, Restle T. Nucleic Acids Res. 2006;34:6561–6573. doi: 10.1093/nar/gkl941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zatsepin TS, Turner JJ, Oretskaya TS, Gait MJ. Curr Pharm Des. 2005;11:3639–3654. doi: 10.2174/138161205774580769. [DOI] [PubMed] [Google Scholar]
- 9.Tung CH, Weissleder R. Adv Drug Deliv Rev. 2003;55:281–294. doi: 10.1016/s0169-409x(02)00183-7. [DOI] [PubMed] [Google Scholar]
- 10.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 12.Futaki S, Goto S, Suzuki T, Nakase I, Sugiura Y. Curr Protein Pept Sci. 2003;4:87–96. doi: 10.2174/1389203033487261. [DOI] [PubMed] [Google Scholar]
- 13.Nori A, Jensen KD, Tijerina M, Kopeckova P, Kopecek J. Bioconjug Chem. 2003;14:44–50. doi: 10.1021/bc0255900. [DOI] [PubMed] [Google Scholar]
- 14.Zhao M, Kircher MF, Josephson L, Weissleder R. Bioconjug Chem. 2002;13:840–844. doi: 10.1021/bc0255236. [DOI] [PubMed] [Google Scholar]
- 15.O’Donnell JM, Alpert NM, White LT, Lewandowski ED. Biophys J. 2002;82:11–18. doi: 10.1016/S0006-3495(02)75369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Carneado J, Kogan MJ, Van Mau N, Pujals S, Lopez-Iglesias C, Heitz F, Giralt E. J Pept Res. 2005;65:580–590. doi: 10.1111/j.1399-3011.2005.00253.x. [DOI] [PubMed] [Google Scholar]
- 17.Futaki S, Ohashi W, Suzuki T, Niwa M, Tanaka S, Ueda K, Harashima H, Sugiura Y. Bioconjug Chem. 2001;12:1005–1011. doi: 10.1021/bc015508l. [DOI] [PubMed] [Google Scholar]
- 18.Khalil IA, Futaki S, Niwa M, Baba Y, Kaji N, Kamiya H, Harashima H. Gene Ther. 2004;11:636–644. doi: 10.1038/sj.gt.3302128. [DOI] [PubMed] [Google Scholar]
- 19.Koppelhus U, Shiraishi T, Zachar V, Pankratova S, Nielsen PE. Bioconjug Chem. 2008;19:1526–1534. doi: 10.1021/bc800068h. [DOI] [PubMed] [Google Scholar]
- 20.Niidome T, Urakawa M, Takaji K, Matsuo Y, Ohmori N, Wada A, Hirayama T, Aoyagi H. J Pept Res. 1999;54:361–367. doi: 10.1034/j.1399-3011.1999.00122.x. [DOI] [PubMed] [Google Scholar]
- 21.Pham W, Kircher MF, Weissleder R, Tung CH. Chembiochem. 2004;5:1148–1151. doi: 10.1002/cbic.200400063. [DOI] [PubMed] [Google Scholar]
- 22.Vijayaraghavan S, Goueli SA, Davey MP, Carr DW. J Biol Chem. 1997;272:4747–4752. doi: 10.1074/jbc.272.8.4747. [DOI] [PubMed] [Google Scholar]
- 23.Pham W, Zhao BQ, Lo EH, Medarova Z, Rosen B, Moore A. NeuroImage. 2005;28:287–292. doi: 10.1016/j.neuroimage.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Pham W, Xie J, Gore JC. Neoplasia (New York, NY. 2007;9:1130–1137. doi: 10.1593/neo.07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 26.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 27.Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y, Takehashi M, Tanaka S, Ueda K, Simpson JC, Jones AT, Sugiura Y, Futaki S. Mol Ther. 2004;10:1011–1022. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2002;277:2437–2443. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 29.Resh MD. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 30.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Science (New York, NY. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 31.Peitzsch RM, McLaughlin S. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 32.Silvius JR, l’Heureux F. Biochemistry. 1994;33:3014–3022. doi: 10.1021/bi00176a034. [DOI] [PubMed] [Google Scholar]
- 33.Mae M, El Andaloussi S, Lundin P, Oskolkov N, Johansson HJ, Guterstam P, Langel U. J Control Release. 2009;134:221–227. doi: 10.1016/j.jconrel.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Lehto T, Abes R, Oskolkov N, Suhorutsenko J, Copolovici DM, Mager I, Viola JR, Simonson OE, Ezzat K, Guterstam P, Eriste E, Smith CI, Lebleu B, Samir El A, Langel U. J Control Release. 2010;141:42–51. doi: 10.1016/j.jconrel.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 35.Puckett CA, Barton JK. J Am Chem Soc. 2009;131:8738–8739. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. J Biol Chem. 2003;278:31192–31201. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- 37.Potocky TB, Menon AK, Gellman SH. J Biol Chem. 2003;278:50188–50194. doi: 10.1074/jbc.M308719200. [DOI] [PubMed] [Google Scholar]
- 38.Fischer R, Kohler K, Fotin-Mleczek M, Brock R. J Biol Chem. 2004;279:12625–12635. doi: 10.1074/jbc.M311461200. [DOI] [PubMed] [Google Scholar]
- 39.Fretz MM, Penning NA, Al-Taei S, Futaki S, Takeuchi T, Nakase I, Storm G, Jones AT. Biochem J. 2007;403:335–342. doi: 10.1042/BJ20061808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Andaloussi S, Johansson HJ, Holm T, Langel U. Mol Ther. 2007;15:1820–1826. doi: 10.1038/sj.mt.6300255. [DOI] [PubMed] [Google Scholar]